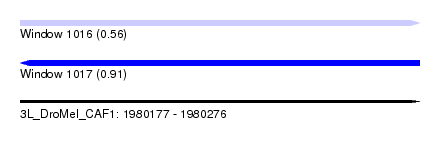
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,980,177 – 1,980,276 |
| Length | 99 |
| Max. P | 0.912274 |

| Location | 1,980,177 – 1,980,276 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.35 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -5.73 |
| Energy contribution | -6.09 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
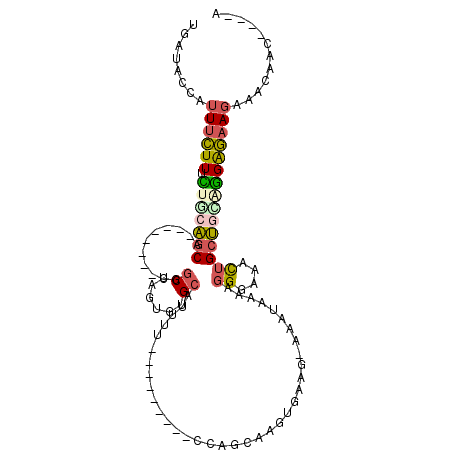
>3L_DroMel_CAF1 1980177 99 + 23771897 U----GUUGUUUCUUCUCCAACAGCAGUUUCCCUUUUAUUU-CUUCACUUGCUUGACACAAAAAAAAAUGCACACUAGCCG--------UGCUGCAGAAAGAAAUAGUAUCA (----((((((........))))))).........((((((-((((..(((.......))).......(((((((.....)--------)).))))).)))))))))..... ( -16.90) >DroSec_CAF1 31189 92 + 1 U----GUUGUUUCUUCUCCUGCAGCAGUUUCCCUUUUAUUU-CUUCACUCGCUGG-------AAGAAAUGCACACUAGCCG--------UGCUGCAGAAAGAAAUGGUAUCA (----(..(((((((...(((((((((((.......(((((-((((.(.....))-------))))))))......)))..--------)))))))).)))))))..))... ( -28.22) >DroSim_CAF1 45379 92 + 1 U----GUUGUUUCUUCUCCUGCAGCAGUUUCCCUUUUAUUU-CUUCACUUGCUGG-------AAGAAAUGCACACUGGCCG--------UGCUGCAGAAAGAAAUGGUAUCA (----(..(((((((...(((((((((((.......(((((-((((.(.....))-------))))))))......)))..--------)))))))).)))))))..))... ( -28.12) >DroEre_CAF1 36976 95 + 1 UGUCUGUUAUUUCUUCUCCACUAGCAGUUUCCCUUUUAUAUAGUAUACUUGCUGA---------AAAAUGCACACUAGCAG--------UGCUGCAAAAAGAAAUAGUAUCA ......(((((((((......(((((((....((.......))...)).))))).---------....(((((((.....)--------)).))))..)))))))))..... ( -16.60) >DroYak_CAF1 31989 98 + 1 G----GUUAUUUCUUCUCCCCUAGCAGUUUCCCUUUUAUUU-CUUUACUUGCUGA---------AAAAUGCACACUUGCAGUGCUACAGUGCUGCAAAAAGAAAUAGUAUCA (----(..(((.((........)).)))..))...((((((-((((...(((...---------.....)))...((((((..(....)..))))))))))))))))..... ( -23.40) >DroAna_CAF1 29155 91 + 1 U----CCCGCCCCUCGCCUCUCGCCCAUCGCCCUCUGACUA-UUGCUAUUGCUAU---------CAGUUUCCCUUUCGCAG-------GAGUCGAAAAAACUAUGGGUAUCA .----...((((.(((.(((..((..........((((...-..((....))..)---------)))..........))..-------))).))).........)))).... ( -16.95) >consensus U____GUUGUUUCUUCUCCACCAGCAGUUUCCCUUUUAUUU_CUUCACUUGCUGG_________AAAAUGCACACUAGCAG________UGCUGCAAAAAGAAAUAGUAUCA ......(((((((((......((((((.....................))))))..............((((((...............)).))))..)))))))))..... ( -5.73 = -6.09 + 0.37)

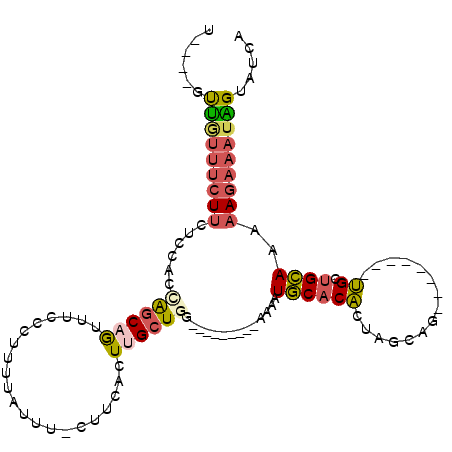
| Location | 1,980,177 – 1,980,276 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.35 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -12.33 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1980177 99 - 23771897 UGAUACUAUUUCUUUCUGCAGCA--------CGGCUAGUGUGCAUUUUUUUUUGUGUCAAGCAAGUGAAG-AAAUAAAAGGGAAACUGCUGUUGGAGAAGAAACAAC----A ........(((((((..((((((--------.(((......)).(((((((((((.(((......)))..-..))))))))))).))))))).))))))).......----. ( -21.40) >DroSec_CAF1 31189 92 - 1 UGAUACCAUUUCUUUCUGCAGCA--------CGGCUAGUGUGCAUUUCUU-------CCAGCGAGUGAAG-AAAUAAAAGGGAAACUGCUGCAGGAGAAGAAACAAC----A ........((((((.(((((((.--------..((......))(((((((-------((.....).))))-))))....((....))))))))))))))).......----. ( -28.20) >DroSim_CAF1 45379 92 - 1 UGAUACCAUUUCUUUCUGCAGCA--------CGGCCAGUGUGCAUUUCUU-------CCAGCAAGUGAAG-AAAUAAAAGGGAAACUGCUGCAGGAGAAGAAACAAC----A ........((((((.((((((((--------.(.((..(....(((((((-------((.....).))))-))))..)..))...))))))))))))))).......----. ( -27.00) >DroEre_CAF1 36976 95 - 1 UGAUACUAUUUCUUUUUGCAGCA--------CUGCUAGUGUGCAUUUU---------UCAGCAAGUAUACUAUAUAAAAGGGAAACUGCUAGUGGAGAAGAAAUAACAGACA ......(((((((((((....((--------((((((((((((.....---------.......)))))))).......((....)))).)))))))))))))))....... ( -22.70) >DroYak_CAF1 31989 98 - 1 UGAUACUAUUUCUUUUUGCAGCACUGUAGCACUGCAAGUGUGCAUUUU---------UCAGCAAGUAAAG-AAAUAAAAGGGAAACUGCUAGGGGAGAAGAAAUAAC----C ......(((((((((.((((.(((((((....))).)))))))).((.---------.((((......(.-...)....((....))))).)..)))))))))))..----. ( -23.30) >DroAna_CAF1 29155 91 - 1 UGAUACCCAUAGUUUUUUCGACUC-------CUGCGAAAGGGAAACUG---------AUAGCAAUAGCAA-UAGUCAGAGGGCGAUGGGCGAGAGGCGAGGGGCGGG----A .....(((((.(((((((.((((.-------.(((....((....)).---------.((....))))).-.))))))))))).)))))(....)((.....))...----. ( -23.60) >consensus UGAUACCAUUUCUUUCUGCAGCA________CGGCUAGUGUGCAUUUU_________CCAGCAAGUGAAG_AAAUAAAAGGGAAACUGCUGCAGGAGAAGAAACAAC____A ........((((((.(((((((...........((......))....................................((....)))))))))))))))............ (-12.33 = -12.62 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:00 2006