
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,707,624 – 17,707,756 |
| Length | 132 |
| Max. P | 0.913268 |

| Location | 17,707,624 – 17,707,719 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17707624 95 + 23771897 UUAAUAUUUGUCUAGGCACAAUUUAUUAUUGUGUUUCGCUCAGCCGUGGCGAGGGUAAUAUUUCAGGUGGAUUGCCA-----UUGCCACUGAAGAAUUUU ..........((((((((((((.....))))))))).........(((((((.((((((...........)))))).-----)))))))...)))..... ( -24.80) >DroSec_CAF1 82624 94 + 1 UCAAUAUUUGUCUAGGCACAAUUUAUUAUUGUGUUUUGCUCAGCCGUGGCGAUGAAAAUAUUUGUG------UGCCUCCUAGGAGGCACUGAAGAAUUUU (((...((..((.(((((((((.....)))))))))......((....))))..)).........(------((((((....))))))))))........ ( -27.50) >DroSim_CAF1 79380 94 + 1 UCAAUAUUUGUCUAGGCACAAUUUAUUAUUGUGUUUCGCUCAGCCGUGGCGAGGAAAAUAUUUGUG------UGCCUCCUAGGAGGCACUGAAGAAUUUU ..(((((((.(((.((((((((.....))))))))(((((.......)))))))))))))))...(------((((((....)))))))........... ( -29.00) >consensus UCAAUAUUUGUCUAGGCACAAUUUAUUAUUGUGUUUCGCUCAGCCGUGGCGAGGAAAAUAUUUGUG______UGCCUCCUAGGAGGCACUGAAGAAUUUU ..(((((((.(((.((((((((.....))))))))(((((.......)))))))))))))))..........((((((....))))))............ (-18.48 = -18.93 + 0.45)



| Location | 17,707,624 – 17,707,719 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -14.91 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17707624 95 - 23771897 AAAAUUCUUCAGUGGCAA-----UGGCAAUCCACCUGAAAUAUUACCCUCGCCACGGCUGAGCGAAACACAAUAAUAAAUUGUGCCUAGACAAAUAUUAA ....(((((((((.(...-----((((.......................))))).)))))).))).((((((.....))))))................ ( -16.30) >DroSec_CAF1 82624 94 - 1 AAAAUUCUUCAGUGCCUCCUAGGAGGCA------CACAAAUAUUUUCAUCGCCACGGCUGAGCAAAACACAAUAAUAAAUUGUGCCUAGACAAAUAUUGA ...........(((((((....))))))------)...(((((((((.((((....)).))......((((((.....))))))....)).))))))).. ( -24.70) >DroSim_CAF1 79380 94 - 1 AAAAUUCUUCAGUGCCUCCUAGGAGGCA------CACAAAUAUUUUCCUCGCCACGGCUGAGCGAAACACAAUAAUAAAUUGUGCCUAGACAAAUAUUGA ...........(((((((....))))))------).(((....(((((((((....)).))).))))((((((.....))))))............))). ( -27.20) >consensus AAAAUUCUUCAGUGCCUCCUAGGAGGCA______CACAAAUAUUUUCCUCGCCACGGCUGAGCGAAACACAAUAAUAAAUUGUGCCUAGACAAAUAUUGA ............((((((....))))))..........((((((...(((((....)).))).....((((((.....))))))........)))))).. (-14.91 = -15.47 + 0.56)

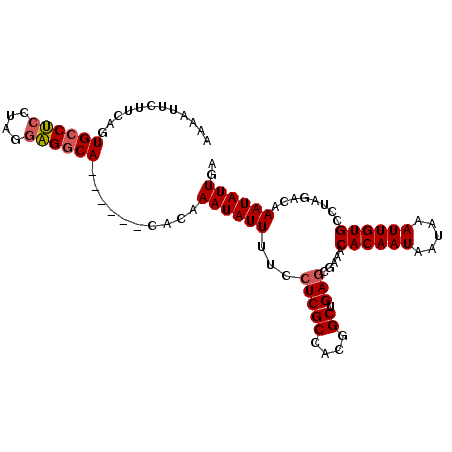

| Location | 17,707,657 – 17,707,756 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -13.51 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17707657 99 - 23771897 UAUAGUAAUGUACAAGCA-UAUUGUGUUUUACGCUGU-AAAAAUUCUUCAGUGGCAA-----UGGCAAUCCACCUGAAAUAUUACCCUCGCCACGGCUGAGCGAAA ....((((..(((((...-..)))))..)))).....-.....(((((((((.(...-----((((.......................))))).)))))).))). ( -19.00) >DroSec_CAF1 82657 95 - 1 CAUAGUAAUGCACUAAUAAAAUUCUGU-----ACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCA------CACAAAUAUUUUCAUCGCCACGGCUGAGCAAAA ........(((................-----...(((((((........(((((((....))))))------).......))))))).((....))...)))... ( -22.66) >DroSim_CAF1 79413 95 - 1 CAUAGUAAUGCACUAAUAAAAUUCUGU-----ACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCA------CACAAAUAUUUUCCUCGCCACGGCUGAGCGAAA (((((((...................)-----))))))............(((((((....))))))------)........(((((((((....)).))).)))) ( -26.21) >consensus CAUAGUAAUGCACUAAUAAAAUUCUGU_____ACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCA______CACAAAUAUUUUCCUCGCCACGGCUGAGCGAAA ((((((.....((............)).....)))))).............((((((....))))))..............((((.(((((....)).))).)))) (-13.51 = -14.07 + 0.56)

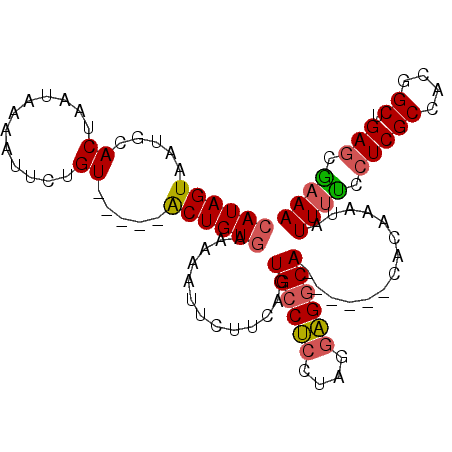
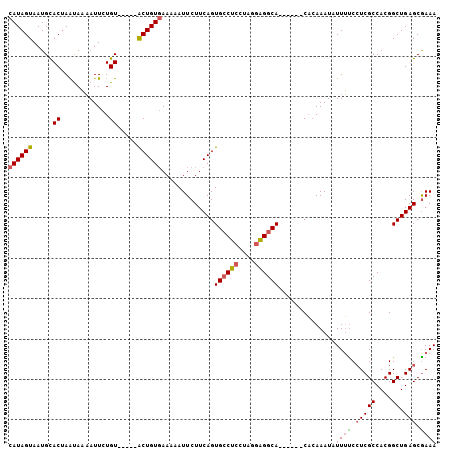
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:29 2006