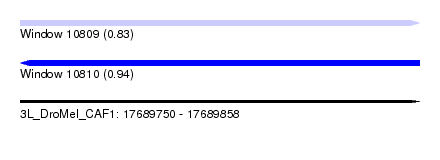
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,689,750 – 17,689,858 |
| Length | 108 |
| Max. P | 0.935483 |

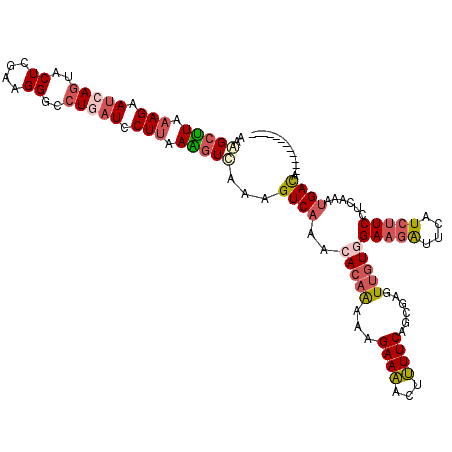
| Location | 17,689,750 – 17,689,858 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17689750 108 + 23771897 ------------UGUCAUUUGAGGAAGAAGAAUCUUCUACAACACGCUGAAAAGUUUUCUUUUUGUGCUUGACUUUAACUUUAAGAAUCAGGCCCUUUGAGAACUGAUUCUUUAAGCUUU ------------.((((.(((.((((((....)))))).)))...(((((((((....))))))).)).)))).....(((.(((((((((..(......)..))))))))).))).... ( -27.70) >DroSec_CAF1 71914 108 + 1 ------------UGUCAUUUGAGGAAGAUGAAUCUUCCACAACCCGCUGAAAAGUUUUCUUUUUGUGUUUGACCUUAACUUUAAGGAUCAGGCCCUUCGAGUACUGAUUCUUUAAGCUUU ------------.((((.(((.((((((....)))))).)))..(((.((((((....)))))))))..)))).....(((.(((((((((..(......)..))))))))).))).... ( -28.20) >DroSim_CAF1 66962 108 + 1 ------------UGUCAUUUGAGGAAGAUGAAUCUUCCACAACCCGUUGAAAAGUUUUCUUUUUGUGUUUGACCUUAACUUUAAGGAUCAGGCCCUUCGAGUACUGAUUCUUUAAGCUUU ------------.((((.(((.((((((....)))))).)))......((((((....)))))).....)))).....(((.(((((((((..(......)..))))))))).))).... ( -26.20) >DroEre_CAF1 61854 108 + 1 ------------UGUCAGUUGAGGAAGACGAAUCUUCCACAACUCGCUGAAAGGUUUUCUUUUUGUGUUUGACUUUGACCUUAAGGAUCAGGCCCUUCGAGUACUGAUUCUUCAAGCUCU ------------.((((((((.((((((....)))))).)))).(((.(((((.....))))).)))..))))...((.((((((((((((..(......)..))))))))).))).)). ( -32.80) >DroYak_CAF1 47997 108 + 1 ------------UAUCAGCUGACGAAGAUGAGUCUUCCACAACUCUCUGAAAGGUUUUCUUUUUGUGUUUGACUUUGACCUUAAGGAUAAGGCCCUUUGAGUACUGAUUCUUUAGGCUUU ------------....((((...(((((.(((((...(((((....((....))........)))))...)))))....(((((((........))))))).......))))).)))).. ( -20.10) >DroAna_CAF1 62118 117 + 1 AAUCCAUCAUAAUGUCUUC---GGACGAAGAGCUCUCUAUCACUCGCUGAAGAGCCUUCUUCCUUUGCUUGACUUUGAUCUUCAGGAUUAGGCCCUUGGAGUAGUUGUUCUUCAGGCCCU (((((........(((...---.)))((((.(((((...(((.....)))))))))))).........................))))).((((..(((((........))))))))).. ( -32.60) >consensus ____________UGUCAUUUGAGGAAGAUGAAUCUUCCACAACUCGCUGAAAAGUUUUCUUUUUGUGUUUGACUUUAACCUUAAGGAUCAGGCCCUUCGAGUACUGAUUCUUUAAGCUUU .............((((.....((((((....))))))..........((((((....)))))).....)))).....(((.(((((((((..(......)..))))))))).))).... (-17.66 = -18.25 + 0.59)

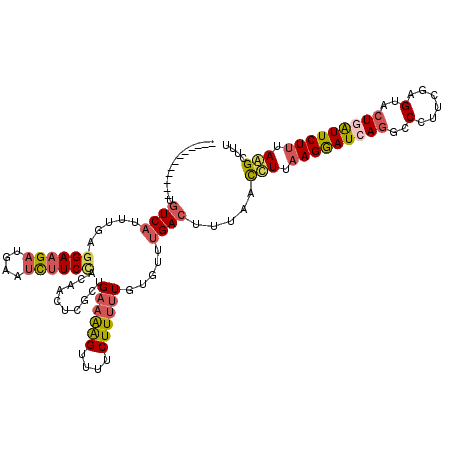

| Location | 17,689,750 – 17,689,858 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -15.35 |
| Energy contribution | -16.52 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17689750 108 - 23771897 AAAGCUUAAAGAAUCAGUUCUCAAAGGGCCUGAUUCUUAAAGUUAAAGUCAAGCACAAAAAGAAAACUUUUCAGCGUGUUGUAGAAGAUUCUUCUUCCUCAAAUGACA------------ ..(((((.(((((((((..((....))..))))))))).)))))...((((.((...(((((....)))))..))...(((..(((((....)))))..))).)))).------------ ( -31.30) >DroSec_CAF1 71914 108 - 1 AAAGCUUAAAGAAUCAGUACUCGAAGGGCCUGAUCCUUAAAGUUAAGGUCAAACACAAAAAGAAAACUUUUCAGCGGGUUGUGGAAGAUUCAUCUUCCUCAAAUGACA------------ ..(((((.(((.(((((..((....))..))))).))).)))))...((((......(((((....))))).......(((.((((((....)))))).))).)))).------------ ( -26.20) >DroSim_CAF1 66962 108 - 1 AAAGCUUAAAGAAUCAGUACUCGAAGGGCCUGAUCCUUAAAGUUAAGGUCAAACACAAAAAGAAAACUUUUCAACGGGUUGUGGAAGAUUCAUCUUCCUCAAAUGACA------------ ..(((((.(((.(((((..((....))..))))).))).)))))...((((......(((((....))))).......(((.((((((....)))))).))).)))).------------ ( -26.20) >DroEre_CAF1 61854 108 - 1 AGAGCUUGAAGAAUCAGUACUCGAAGGGCCUGAUCCUUAAGGUCAAAGUCAAACACAAAAAGAAAACCUUUCAGCGAGUUGUGGAAGAUUCGUCUUCCUCAACUGACA------------ .((.(((((.((.((((..((....))..)))))).))))).))...(((...(.(..((((.....))))..).)(((((.((((((....)))))).)))))))).------------ ( -31.40) >DroYak_CAF1 47997 108 - 1 AAAGCCUAAAGAAUCAGUACUCAAAGGGCCUUAUCCUUAAGGUCAAAGUCAAACACAAAAAGAAAACCUUUCAGAGAGUUGUGGAAGACUCAUCUUCGUCAGCUGAUA------------ ............((((((...(.((((((((((....)))))))..((((...(((((...(((.....)))......)))))...))))...))).)...)))))).------------ ( -23.70) >DroAna_CAF1 62118 117 - 1 AGGGCCUGAAGAACAACUACUCCAAGGGCCUAAUCCUGAAGAUCAAAGUCAAGCAAAGGAAGAAGGCUCUUCAGCGAGUGAUAGAGAGCUCUUCGUCC---GAAGACAUUAUGAUGGAUU .((((((...((........))...))))))(((((....(((....))).......(((.(((((((((..(........)..))))).)))).)))---..............))))) ( -31.60) >consensus AAAGCUUAAAGAAUCAGUACUCGAAGGGCCUGAUCCUUAAAGUCAAAGUCAAACACAAAAAGAAAACUUUUCAGCGAGUUGUGGAAGAUUCAUCUUCCUCAAAUGACA____________ ..(((((.(((.(((((..((....))..))))).))).)))))...((((..(((((...((((...))))......)))))(((((....)))))......))))............. (-15.35 = -16.52 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:27 2006