| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,685,947 – 17,686,037 |
| Length | 90 |
| Max. P | 0.701981 |

| Location | 17,685,947 – 17,686,037 |
|---|---|
| Length | 90 |
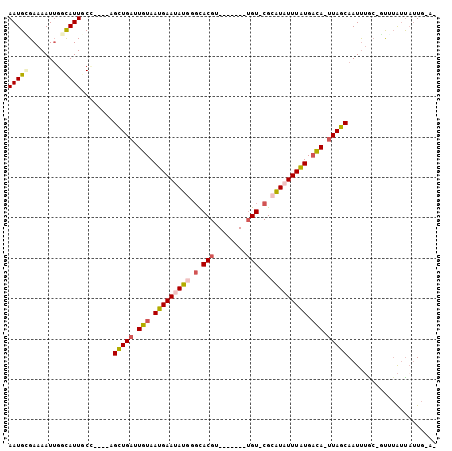
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
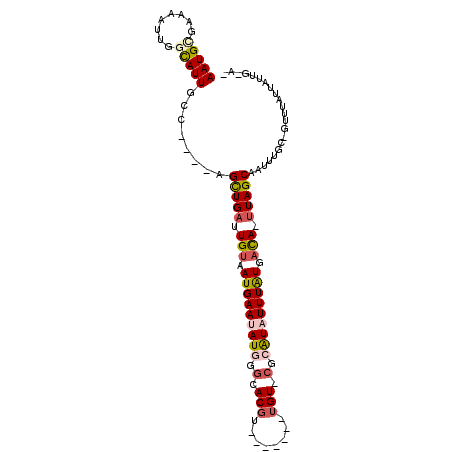
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -14.55 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17685947 90 - 23771897 AAUGCGAAAAUUGGCAUUGCU----AGCUGAUUGUAAUGAAUAUGGCCACGU-------UGU-CGCAUCUUUAUGACA-UUAGCAAUCUAC-GUUUAUUAUUG-A- .......(((((((.((((((----((.((.(..(((.((...((((.....-------.))-))..)).)))..)))-))))))))))).-)))).......-.- ( -14.90) >DroPse_CAF1 60397 94 - 1 AAUGGGAAAAUUGGCAUUACCAGCGAGCUGAUUGUAAUGAAUAUGGGCACGU-------UGU-CGCAUAUUUGUGACA-UUAGCAAUUUGC-CUUUAUUAUUG-A- ...((.(((.((((.....))))...((((((.((.(..(((((((((....-------.))-).))))))..).)))-)))))..))).)-)..........-.- ( -23.40) >DroGri_CAF1 3451 91 - 1 AAUGCGAAAAUGGGUAUUGCA----CGUUGUUUGUAAUGAAUAUAAGCACGU-------GGU-CGCAUAUUUAUGUCA-UUAGCAAUUAGAUGCGAGUCAUUG-A- ........(((((......((----(((.(((((((.....)))))))))))-------).(-(((((.....(((..-...))).....)))))).))))).-.- ( -21.40) >DroWil_CAF1 113613 101 - 1 AAUGUGAAAAUUGGCAUUGCG----AGCUGAUUGUAAUGAAUAUAUGUACGUGUGUGUGUGUACGAGUAUUUAUGAUACUUAGCAAUUUGC-UUGUACUUAUGGCA ...........((.(((((((----(((.((((((......((((..(((....)))..)))).(((((((...))))))).)))))).))-)))))...))).)) ( -25.70) >DroAna_CAF1 58300 90 - 1 AAUGUGAAAAUUGGCAUUGCC----AGCUGAUUGUAAUGAAUAUGGGCACGU-------UGU-CGCAUCUUUAUGACA-UUAGCAAUUUAC-GUUUAUUAUUG-A- ((((((((...((((...)))----)((((((.((.(((((.((((((....-------.))-).))).))))).)))-)))))..)))))-)))........-.- ( -23.40) >DroPer_CAF1 58557 94 - 1 AAUGGGAAAAUUGGCAUUACCAGCGAGCUGAUUGUAAUGAAUAUGGGCACGU-------UGU-CGCAUAUUUGUGACA-UUAGCAAUUUGC-CUUUAUUAUUG-A- ...((.(((.((((.....))))...((((((.((.(..(((((((((....-------.))-).))))))..).)))-)))))..))).)-)..........-.- ( -23.40) >consensus AAUGCGAAAAUUGGCAUUGCC____AGCUGAUUGUAAUGAAUAUGGGCACGU_______UGU_CGCAUAUUUAUGACA_UUAGCAAUUUGC_GUUUAUUAUUG_A_ (((((........)))))........(((((.(((.(((((((((.(.(((........))).).))))))))).))).)))))...................... (-14.55 = -15.33 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:21 2006