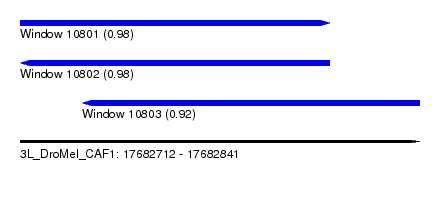
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,682,712 – 17,682,841 |
| Length | 129 |
| Max. P | 0.980778 |

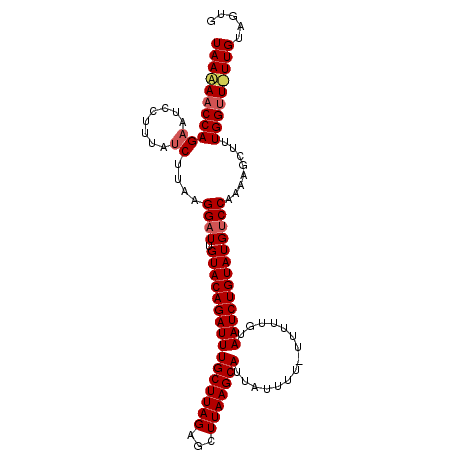
| Location | 17,682,712 – 17,682,812 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -18.78 |
| Energy contribution | -19.89 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17682712 100 + 23771897 UAAAAACCAG------UUCUCUUUUGGAUUGUACAGAUUUGCUUAGAUUUUAAGCAUGUUUUUAUUUUUUUAAUCUGUAUGUCCAAAAGCUGUGGAUUUUGCAGUG (((((.(((.------....(((((((((.((((((((((((((((...)))))))...............))))))))))))))))))...))).)))))..... ( -25.26) >DroSec_CAF1 64873 105 + 1 UAACAACCAGAAUCCUUUAUCUUAAGGAUUGUACAGAUUUGCUUAGAGCUUAAGCAUUAUUUU-UUUUUGUAAUCUGUAUGCCCAAAAGCUUUGGUUGUUGUUGUG (((((((((((((((((......)))))))((((((((((((((((...))))))).......-.......)))))))))...........))))))))))..... ( -29.83) >DroSim_CAF1 59986 102 + 1 UAAAAACCAGAAUCCUUUAUCUCAAGGAUUGUACAGAUUUGCUUAGAGCUUAAGCAUUAUUUU---UUUGUAAUCUGUAUGUCCAAAAGCUUUGGUU-UUGUGGUG ..(((((((((((((((......)))))))((((((((((((((((...))))))).......---.....)))))))))...........))))))-))...... ( -25.30) >consensus UAAAAACCAGAAUCCUUUAUCUUAAGGAUUGUACAGAUUUGCUUAGAGCUUAAGCAUUAUUUU_UUUUUGUAAUCUGUAUGUCCAAAAGCUUUGGUU_UUGUAGUG (((((((((((........))....((((.((((((((((((((((...)))))))...............)))))))))))))........)))))))))..... (-18.78 = -19.89 + 1.11)

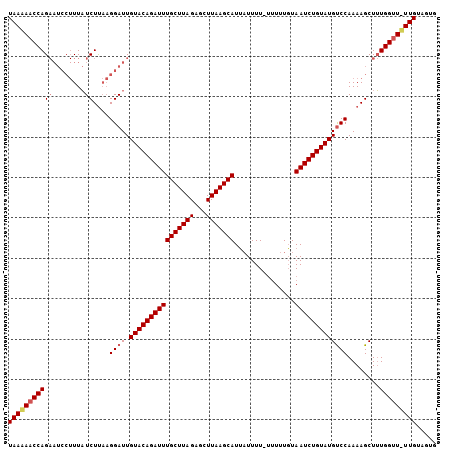
| Location | 17,682,712 – 17,682,812 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17682712 100 - 23771897 CACUGCAAAAUCCACAGCUUUUGGACAUACAGAUUAAAAAAAUAAAAACAUGCUUAAAAUCUAAGCAAAUCUGUACAAUCCAAAAGAGAA------CUGGUUUUUA ......((((.(((...((((((((..((((((((...............((((((.....))))))))))))))...))))))))....------.))).)))). ( -24.06) >DroSec_CAF1 64873 105 - 1 CACAACAACAACCAAAGCUUUUGGGCAUACAGAUUACAAAAA-AAAAUAAUGCUUAAGCUCUAAGCAAAUCUGUACAAUCCUUAAGAUAAAGGAUUCUGGUUGUUA ......((((((((..(((....))).((((((((.......-.......((((((.....)))))))))))))).(((((((......))))))).)))))))). ( -28.53) >DroSim_CAF1 59986 102 - 1 CACCACAA-AACCAAAGCUUUUGGACAUACAGAUUACAAA---AAAAUAAUGCUUAAGCUCUAAGCAAAUCUGUACAAUCCUUGAGAUAAAGGAUUCUGGUUUUUA ......((-(((((.............((((((((.....---.......((((((.....)))))))))))))).(((((((......))))))).))))))).. ( -23.60) >consensus CACAACAA_AACCAAAGCUUUUGGACAUACAGAUUACAAAAA_AAAAUAAUGCUUAAGCUCUAAGCAAAUCUGUACAAUCCUUAAGAUAAAGGAUUCUGGUUUUUA ......((((((((...(((..(((..((((((((...............((((((.....))))))))))))))...)))..)))...........)))))))). (-19.86 = -19.87 + 0.01)

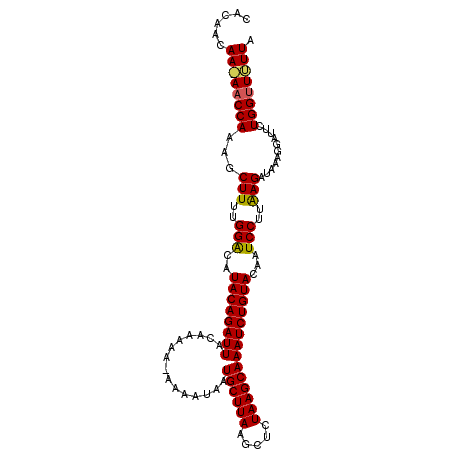
| Location | 17,682,732 – 17,682,841 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.96 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.28 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17682732 109 - 23771897 ACGAAAAAGUCGAAAAAAUUGUAGUUUAGCACUGCAAAAUCCACAGCUUUUGGACAUACAGAUUAAAAAAAUAAAAACAUGCUUAAAAUCUAAGCAAAUCUGUACAAUC .(((.....)))......(((((((.....)))))))..((((.......))))..((((((((...............((((((.....))))))))))))))..... ( -20.16) >DroSec_CAF1 64899 108 - 1 ACGAAAAAGUCGCUAAAAUUGUAGUUUAUCACAACAACAACCAAAGCUUUUGGGCAUACAGAUUACAAAAA-AAAAUAAUGCUUAAGCUCUAAGCAAAUCUGUACAAUC ...........((.....((((.(((......))).))))((((.....)))))).((((((((.......-.......((((((.....))))))))))))))..... ( -17.83) >DroSim_CAF1 60012 105 - 1 ACGAAAAAGUCGCUAAAAUUGUAGUUUAUCACCACAA-AACCAAAGCUUUUGGACAUACAGAUUACAAA---AAAAUAAUGCUUAAGCUCUAAGCAAAUCUGUACAAUC ........(((((((......)))).........(((-((.......)))))))).((((((((.....---.......((((((.....))))))))))))))..... ( -16.20) >consensus ACGAAAAAGUCGCUAAAAUUGUAGUUUAUCACAACAA_AACCAAAGCUUUUGGACAUACAGAUUACAAAAA_AAAAUAAUGCUUAAGCUCUAAGCAAAUCUGUACAAUC ....((((((........((((.((.....)).))))........)))))).....((((((((...............((((((.....))))))))))))))..... (-15.50 = -15.28 + -0.22)

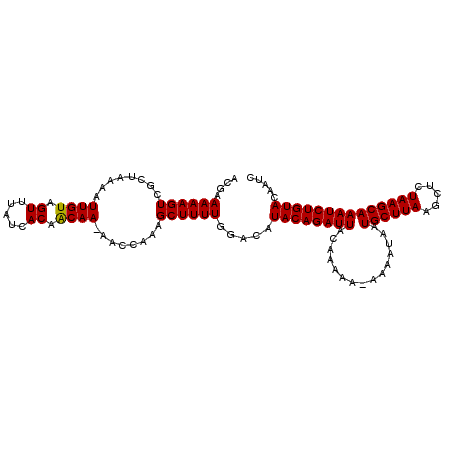
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:20 2006