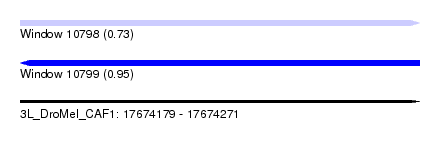
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,674,179 – 17,674,271 |
| Length | 92 |
| Max. P | 0.954900 |

| Location | 17,674,179 – 17,674,271 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
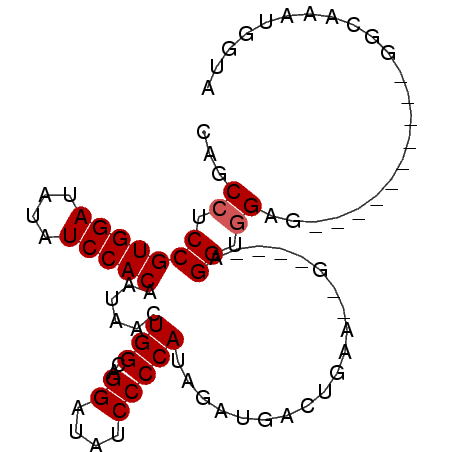
>3L_DroMel_CAF1 17674179 92 + 23771897 CAGCCUCCGUGGAUAUAUCCACAGUAACUGGCUGGAUAUCCCCAUAAAUGACGAAAGGGGGGGGGGUGGGGGGUGAGUGGAGUCAAAUGGUA ..(.(((((((((....)))))....(((.(((...(((((((.....(..(....)..).)))))))...))).))))))).)........ ( -32.00) >DroSim_CAF1 51356 78 + 1 CAGCUUCCGUGGAUAUAUCCACAAUAACUGGCAGGAUAUCCCCAUAGAUGACUGAAUGG----GGGUUGGG----------GGCAAAUGGUA ..(((((((((((....)))))...............((((((((..........))))----)))).)))----------)))........ ( -26.30) >DroEre_CAF1 46674 75 + 1 CAGCCUCCGUGGAUAUAUCCACAAUAACUGGCAGGAUAUCCCCAU-GAUGACUGCA--G----AGGUGGAG----------GGCAAAUGGGA ...((((((((((....)))))....(((.((((..((((.....-)))).)))).--.----.)))))))----------).......... ( -25.50) >DroYak_CAF1 32299 76 + 1 CAGCCUCCGUGGAUAUAUCCACAAUAACUGGCAGGAUAUCCCCAUAGAUGACUGCA--A----AGGUGGAG----------GGUAAAUGGUA ...((((((((((....)))))....(((.((((..((((......)))).)))).--.----.)))))))----------).......... ( -25.90) >consensus CAGCCUCCGUGGAUAUAUCCACAAUAACUGGCAGGAUAUCCCCAUAGAUGACUGAA__G____AGGUGGAG__________GGCAAAUGGUA ...((.(((((((....)))))......(((..((....)))))....................)).))....................... (-13.38 = -13.62 + 0.25)


| Location | 17,674,179 – 17,674,271 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.05 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
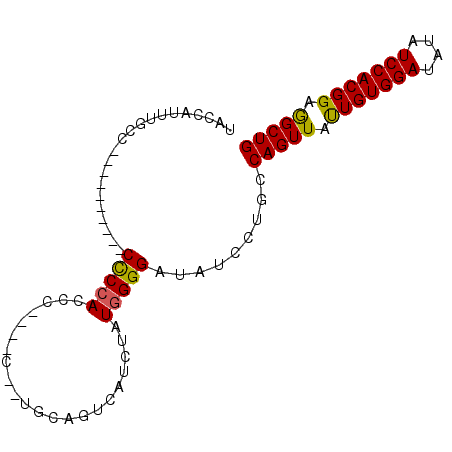
>3L_DroMel_CAF1 17674179 92 - 23771897 UACCAUUUGACUCCACUCACCCCCCACCCCCCCCCCUUUCGUCAUUUAUGGGGAUAUCCAGCCAGUUACUGUGGAUAUAUCCACGGAGGCUG ..((((.((((.............................))))...))))((....))...((((..(((((((....)))))))..)))) ( -22.45) >DroSim_CAF1 51356 78 - 1 UACCAUUUGCC----------CCCAACCC----CCAUUCAGUCAUCUAUGGGGAUAUCCUGCCAGUUAUUGUGGAUAUAUCCACGGAAGCUG ...........----------......((----((((..........)))))).........(((((.(((((((....))))))).))))) ( -21.30) >DroEre_CAF1 46674 75 - 1 UCCCAUUUGCC----------CUCCACCU----C--UGCAGUCAUC-AUGGGGAUAUCCUGCCAGUUAUUGUGGAUAUAUCCACGGAGGCUG ........(((----------.(((....----(--(((((..(((-.....)))...))).))).....(((((....))))))))))).. ( -20.70) >DroYak_CAF1 32299 76 - 1 UACCAUUUACC----------CUCCACCU----U--UGCAGUCAUCUAUGGGGAUAUCCUGCCAGUUAUUGUGGAUAUAUCCACGGAGGCUG ..........(----------((((....----.--.((((..((((....))))...))))........(((((....))))))))))... ( -21.60) >consensus UACCAUUUGCC__________CCCCACCC____C__UGCAGUCAUCUAUGGGGAUAUCCUGCCAGUUAUUGUGGAUAUAUCCACGGAGGCUG .....................(((((......................))))).........(((((.(((((((....))))))).))))) (-16.42 = -16.05 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:16 2006