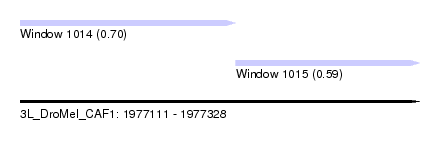
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,977,111 – 1,977,328 |
| Length | 217 |
| Max. P | 0.704833 |

| Location | 1,977,111 – 1,977,228 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1977111 117 + 23771897 --GACUUGGGACUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUU-UGGUCAAUUUGACAAAAUUGUUUGAGCUUAGCACAGUAUGGUGGUGGUCUUCAAGGUGAG --..(((((((((......(((.(((((((((..((((((.......(((((....)))-))(((.....))).....)))))).)))).))))).)))......)))).)))))..... ( -30.00) >DroPse_CAF1 36251 103 + 1 ----CCAGGGACUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUUUUGGUCAAUUUGACAAAAUUGUUUGAGCUUAGUGCCA-------------CGAAAGGUGAG ----...((.(((((.(((((..((((.....)))).))))).))).(((((((((((((((.((.....))))))))...))))))))))).)).-------------(....)..... ( -24.30) >DroWil_CAF1 43141 109 + 1 CUGGCCCAGGUCUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUU-UGGUCAAUUUGACAAAAUUGUUUGAGCUUAGCGAAAAGCUUUCGUUUUCU---------- ..(((....)))(((.(((((..((((.....)))).))))).)))....(((.(((((-..(.((((((....)))))).)..))))))))(((((.......))))).---------- ( -23.00) >DroYak_CAF1 28902 104 + 1 --GACUUGGGACUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUU-UGGUCAAUUUGACAAAAUUGUUUGAGCUUAGCCAAG-------------UUUAAGGUGAG --(((((((...(((.(((((..((((.....)))).))))).))).((((((((((((-(.(((.....))).))))...))))))))).)))))-------------))......... ( -25.80) >DroAna_CAF1 24564 96 + 1 --GACUAUGGACUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUU-UGGUCAAUUUGACAAAAUUGUUUGAGCUUAGCGAAAUAU--------------------- --..........(((.(((((..((((.....)))).))))).)))....(((.(((((-..(.((((((....)))))).)..)))))))).......--------------------- ( -19.60) >DroPer_CAF1 35780 103 + 1 ----CCAGGGACUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUUUUGGUCAAUUUGACAAAAUUGUUUGAGCUUAGUGCCA-------------CGAAAGGUGAG ----...((.(((((.(((((..((((.....)))).))))).))).(((((((((((((((.((.....))))))))...))))))))))).)).-------------(....)..... ( -24.30) >consensus __GACCAGGGACUUAAUGUGUAAUGUGUAAGCCGCAAACAUAAUAAAUAAGCUUAAGUU_UGGUCAAUUUGACAAAAUUGUUUGAGCUUAGCGAAA_____________U__AAGGUGAG ............(((.(((((..((((.....)))).))))).))).((((((((((.....(((.....))).......)))))))))).............................. (-16.80 = -16.80 + 0.00)

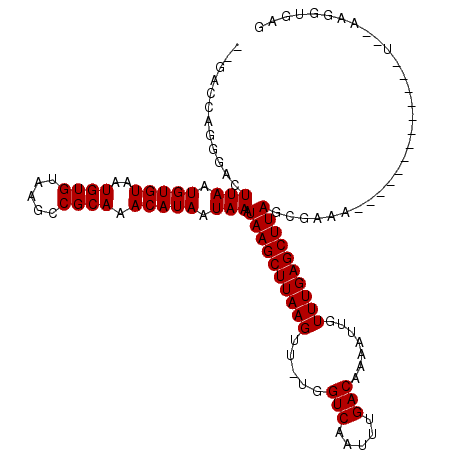
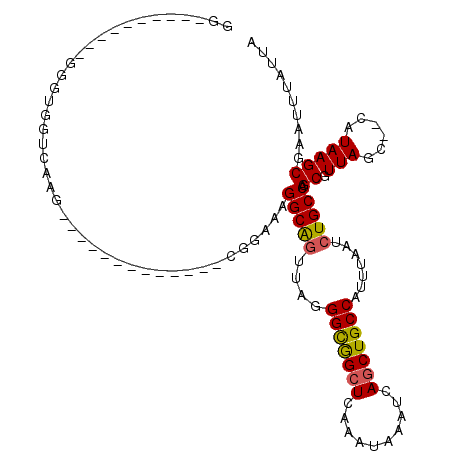
| Location | 1,977,228 – 1,977,328 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1977228 100 + 23771897 GGUGGUG--GAGGGGUGGUCAAG-------------CGGAAGGGCAGUUAGGGUGGCUCAAAUAAAUCAGCUGCCAUUUAAGCUGCCAGCGUUAGCCCCAUAAGCGAAUUUAUUA .......--..(((((......(-------------(.....(((((((..((((((...............))))))..))))))).))....)))))................ ( -32.66) >DroSec_CAF1 28274 100 + 1 GAUGAGGAUGAGGGGCGGUCCAG-------------CGGAAGGGCAGUUAGGGUGGCUCAAAUAAAUCAGCUGCCAUUUAAUCUGCCAGCGUUAGC--CAUAAGCGAAUUUAUCA ((((((..((...(((......(-------------(.....((((((((.((..(((..........)))..))...))).))))).))....))--).....))..)))))). ( -29.90) >DroSim_CAF1 42442 100 + 1 GAUGGGGAUGAGGGGCGGUCAAG-------------CGGAAGGGCAGUUAGGGUGGCUCAAAUAAAUCAGCUGCCAUUUAAUCUGCCAGCGUUAGC--CAUAAGCGAAUUUAUUA .((((.((((...(((((.....-------------(....)((((((...(((...........))).)))))).......)))))..))))..)--))).............. ( -27.40) >DroEre_CAF1 33866 102 + 1 GG----------GUGUGGUCAAGUUGAAUGGGGAG-GGGAGAGGCAGUUAGGGCGGCUCAAAUAAAUCAGCUGCCAUUUAAUCUGCCAGCGUUAGC--CAUAAGCGAAUUUAUUA ..----------..........(((..((((..((-.(....((((((((.(((((((..........)))))))...))).)))))..).))..)--))).))).......... ( -25.40) >DroYak_CAF1 29006 101 + 1 GG----------G--UGGUCAAGUUGGCUGAGGCGGGGGAAAGGCAGUUAGGGCGGCUCAAAUAAAUCAGCUGCCAUUUAAUCUGCCAGCGUUAGC--CAUAAGCGAAUUUAUUA ..----------.--.....(((((.(((..(((..(.(...((((((((.(((((((..........)))))))...))).)))))..).)..))--)...))).))))).... ( -33.90) >DroPer_CAF1 35883 75 + 1 G--------------------------------------AGAGGCGUGCAGGGCAGAUCCUAUAAAUCAGCUGCCAUUUAAUCUGCCAGCGUUAGC--CAUAAGCGAAUUUAUUA .--------------------------------------...(((.(((..(((((((....(((((........)))))))))))).)))...))--)................ ( -18.00) >consensus GG__________GGGUGGUCAAG_____________CGGAAAGGCAGUUAGGGCGGCUCAAAUAAAUCAGCUGCCAUUUAAUCUGCCAGCGUUAGC__CAUAAGCGAAUUUAUUA ..........................................(((((....(((((((..........))))))).......))))).((.(((......))))).......... (-18.44 = -18.25 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:58 2006