
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,658,149 – 17,658,249 |
| Length | 100 |
| Max. P | 0.658509 |

| Location | 17,658,149 – 17,658,249 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -19.41 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17658149 100 + 23771897 AGACUGGGGUGGGAGGGCGUUUUUUACUCGCGCUUUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCAGCGGAAAAAUCCGCAUCGCCCAGGGGCG---A-AC ..(((..((((((((.(((.........))).)))))))).)))......(((............))).(((((....))))).(((((....))))---)-.. ( -31.60) >DroSec_CAF1 35023 96 + 1 GGACUGGGUGGGAUGGGCGU----AGCUCGCGCUUUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCAGCGGAAAAAUCCGCAUCGCCCAGGGGCG---A-AC ...(((((((((((..(.((----(((((((((((.....))))..))))))))).)...)))......(((((....))))).)))))))).....---.-.. ( -39.40) >DroSim_CAF1 35113 96 + 1 AGACUGGGUGGGAUGGGCGU----AGCUCGCGCUUUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCAGCGGAAAAAUCCGCAUCGCCCAGGGGCG---A-AC ...(((((((((((..(.((----(((((((((((.....))))..))))))))).)...)))......(((((....))))).)))))))).....---.-.. ( -39.40) >DroEre_CAF1 31282 93 + 1 AGAAUGGG-GGAA--AGCGC----AGCUCGCGCUUUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCCGUGGAAAAAUCCGCAUCGCCAAGGGGCG---A-AC .((((((.-.(((--(((((----.....)))))))).(((......))).))))))............(((((....))))).(((((....))))---)-.. ( -29.20) >DroYak_CAF1 16312 94 + 1 AGAAUGGGGGGAA--AGCGC----AGCUCGCGCUCUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCAGCGGAAAAAUCCGCAUCGCCAAGGGGCG---A-AU .((((((......--(((((----.....)))))((((((.....))))))))))))............(((((....))))).(((((....))))---)-.. ( -31.20) >DroAna_CAF1 32905 97 + 1 AG---GGGAGAAAAAAGCCA----AGCUUGAGCAUUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCAGCAGAAAAAUCCACAGCCCCCAAGUGGGUUGAGGC ..---...........(((.----.(((....(((((((...))))))).(((............))))))...........((((((......)))))).))) ( -19.70) >consensus AGACUGGGGGGAA_GAGCGU____AGCUCGCGCUUUUAUUAAGUAAGUGAGCUAUUCAAAAUCAAAGCAGCGGAAAAAUCCGCAUCGCCCAGGGGCG___A_AC ........................(((((((((((.....))))..)))))))................(((((....)))))..((((....))))....... (-19.41 = -20.22 + 0.81)

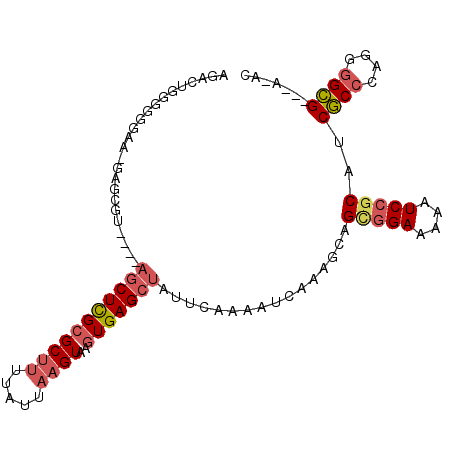

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:09 2006