
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,627,172 – 17,627,308 |
| Length | 136 |
| Max. P | 0.978815 |

| Location | 17,627,172 – 17,627,274 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -17.18 |
| Energy contribution | -16.06 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17627172 102 - 23771897 GGUGUUAUGGAUGACCUGGUGGACUCCAGUGACCUGGACGAGGAAGUGCGCAAUUUCUUUUUUUAGGCAGCC---AG-CAAGUCAUUUUU------GUCGUUAACACAACUG .((((((.(((((((.((.(((((....))(.((((((.((((((((.....)))))))).)))))))..))---).-)).)))))))..------.....))))))..... ( -30.60) >DroPse_CAF1 15313 81 - 1 GGGGUGAUGGAUGAUCUGGUUGACUCCAGCGACCUGGACGAGGAGGUGCGAAAUUUCUUCCUCUAGGUAUAU---CGCCAUGU-----U----------------------- ..(((((((......((((......))))..(((((((.((((((((.....)))))))).))))))).)))---))))....-----.----------------------- ( -31.90) >DroYak_CAF1 8749 108 - 1 GGGGUAAUGGAUGACUUGGUGGACUCCAGUGACCUGGACGAGGAAGUGCGCAAUUUCUUUUUUUAGGCAGGC---AG-CAUGUAAUUAUUGUCAUUGUCGUUAACACAAGUG .............(((((((.(((..((((((((((((.((((((((.....)))))))).)))))((....---.)-)...........)))))))..))).)).))))). ( -29.10) >DroMoj_CAF1 5511 84 - 1 AGUGUUAUAGAUGAACUUGUUGACUCCAGCGAUUUGGAUGAGGAAGUGCGCAAGUUUUUCUUCUAAGUCAAAUCAAGCCAAGU-----U----------------------- .............(((((((((....))))(((((((..(((((((.((....)).)))))))....))))))).....))))-----)----------------------- ( -23.10) >DroPer_CAF1 6821 81 - 1 GGGGUGAUGGAUGAUCUGGUUGACUCCAGCGACCUGGACGAGGAGGUGCGAAAUUUCUUCCUCUAGGUAUAU---CGCCAUGU-----U----------------------- ..(((((((......((((......))))..(((((((.((((((((.....)))))))).))))))).)))---))))....-----.----------------------- ( -31.90) >consensus GGGGUGAUGGAUGACCUGGUUGACUCCAGCGACCUGGACGAGGAAGUGCGCAAUUUCUUCUUCUAGGUAGAC___AGCCAUGU_____U_______________________ ...............((((......))))...((((((.((((((((.....)))))))).))))))............................................. (-17.18 = -16.06 + -1.12)

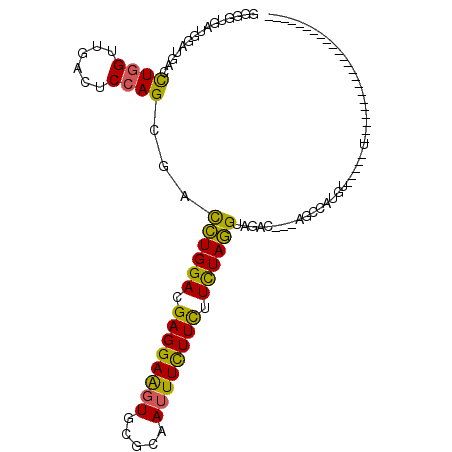
| Location | 17,627,202 – 17,627,308 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.50 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -17.12 |
| Energy contribution | -14.18 |
| Covariance contribution | -2.94 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17627202 106 - 23771897 AAAAGAU------UUGGAAGUUGUGGAACUUCUGGUGUCCGGUGUUAUGGAUGACCUGGUGGACUCCAGUGACCUGGACGAGGAAGUGCGCAAUUUCUUUUUUUAGGCAGCC-------- .(((((.------..((((((((((..((((((..(((((((.((((((((.(.((....)).))))).))))))))))).)))))).)))))))))).)))))........-------- ( -41.10) >DroVir_CAF1 5246 103 - 1 GAAGGAC------UUGGAGGUGGUUGAGCUACUUGUGCCGGGUGUCAUAGAUGAUCUUGUCGACUCCGCCGACUUGGAUGAGGAGGUACGCAAGUUUUUCUUCUAAGUC----------- (((((((------(((.((((((.....))))))(((((.((((.....(((......))).....))))..(((....)))..))))).)))))))))).........----------- ( -31.20) >DroGri_CAF1 5069 114 - 1 AAAGGAU------UUGGAGGUGAUUGAGCUGCUUGUGCCUGGUGUUAUGGAUGAUCUGGUCGAAUCCACCGACUUGGAUGAGGAAGUGCGCAAGUUUUUCUACUAAGUCUAAGUCUAAGU ...((((------((((((((.(((.(((.(((.......))))))...))).))))..))))))))...(((((((((..(((((.((....)).))))).....)))))))))..... ( -33.40) >DroYak_CAF1 8785 106 - 1 AAAAGAU------UUGGAAGUUGUGGAACUUCUGGUGUCCGGGGUAAUGGAUGACUUGGUGGACUCCAGUGACCUGGACGAGGAAGUGCGCAAUUUCUUUUUUUAGGCAGGC-------- .(((((.------..((((((((((..((((((..((((((((....((((.(.((....)).)))))....)))))))).)))))).)))))))))).)))))........-------- ( -39.90) >DroMoj_CAF1 5522 107 - 1 AAAGGAU------UUGGAGGUGAUUGAGCUGCUUGUGCCGAGUGUUAUAGAUGAACUUGUUGACUCCAGCGAUUUGGAUGAGGAAGUGCGCAAGUUUUUCUUCUAAGUCAAA-------U ....(((------(((((((.((.......(((((((((...(.((((...((((.((((((....)))))))))).)))).)..).)))))))).)).))))))))))...-------. ( -28.60) >DroPer_CAF1 6830 112 - 1 GAAGGAGCUGGAGCUGGAGGCGGUGGAACUGCUCGUACCUGGGGUGAUGGAUGAUCUGGUUGACUCCAGCGACCUGGACGAGGAGGUGCGAAAUUUCUUCCUCUAGGUAUAU-------- ......((((((((..((.((((.....))))((((.((.........))))))))..)....))))))).(((((((.((((((((.....)))))))).)))))))....-------- ( -42.00) >consensus AAAGGAU______UUGGAGGUGGUGGAACUGCUUGUGCCCGGUGUUAUGGAUGAUCUGGUCGACUCCAGCGACCUGGACGAGGAAGUGCGCAAGUUCUUCUUCUAAGUAGAA________ ..............((((..(((....((.....))..((((.((((....)))))))))))..))))...(((((((.((((((.(.....).)))))).)))))))............ (-17.12 = -14.18 + -2.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:02 2006