
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,620,456 – 17,620,556 |
| Length | 100 |
| Max. P | 0.559047 |

| Location | 17,620,456 – 17,620,556 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -21.87 |
| Energy contribution | -21.73 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17620456 100 - 23771897 ACACCUCCAGAAUUGCCUUGGCAUAUGUCCGCUGUGCCGCCGGCUGCUGAUCCAAAGUGCAGCAUUGCUUACACUGAGGCCUGAAA-UG---UUAAAAAUGUUU .......(((....(((((((((((.(....)))))))((..(((((...........)))))...)).......))))))))...-..---............ ( -25.80) >DroEre_CAF1 453 100 - 1 ACACCUCCAGAAUGGCCUUGGCAUAGGUACGCUGUGCCGCCGGCUGUUGAUCCAAAGUGCAGCAUUGCUUGCACUGAGGUCUGAAA-UA---UAAAAAAGGCUU ..(((((...(((((((.((((((((.....))))))))..))))))).......((((((((...)).)))))))))))......-..---............ ( -33.40) >DroYak_CAF1 1789 100 - 1 ACACCUCCAGAAUGGCCUUGGCAUAUGUACGCUGUGCCGCCGGCUGUUGAUCCAGAGUGCAGCAUUGCUUGCACUGAGGUCUGAAA-UA---GUACCAAUGUUU ..(((((...(((((((.(((((((.(....))))))))..))))))).......((((((((...)).)))))))))))......-..---............ ( -30.50) >DroMoj_CAF1 163 94 - 1 AAACCUCAAGAAUCGCCUUGGCAUAGGUGCGCUGCGCCGCUGGCUGCUGAUCCUGCGUGCAGCAUUGCUUGCAAUGAGGUCUAAGG-GAUAG--A-------UU ..((((((.....(((((......))))).(((((((.((.((........)).)))))))))...........))))))......-.....--.-------.. ( -29.90) >DroAna_CAF1 154 101 - 1 AGACCUCCAAAAUAGCCUUGGCAUAGGUCCUCUGCGCUGCUGGCUGCUGAUCCAAAGUGCAGCACUGUUUGCAAUGGGGCCUAAGAACA---CAAUAUAAGUAU ..............((....)).((((((((.((((..((..(((((...........)))))...)).))))..))))))))......---............ ( -28.10) >DroPer_CAF1 154 100 - 1 ACACCUCCAGAAUGGCCUUGGCAUAGGUGCGCUGUGCCGCCGGCUGCUGGUCCAGAGUGCAGCACUGCUUGCACGGGGUCCUGUGG-AA---UGGAGGAUGUUA (((((((((...((((...(((((((.....))))))))))).(..(.((.((...(((((((...)).)))))..)).)).)..)-..---)))))).))).. ( -44.40) >consensus ACACCUCCAGAAUGGCCUUGGCAUAGGUACGCUGUGCCGCCGGCUGCUGAUCCAAAGUGCAGCAUUGCUUGCACUGAGGCCUGAAA_UA___UAAAAAAUGUUU ..............((((((((((((.....)))))))((..(((((...........)))))...)).......)))))........................ (-21.87 = -21.73 + -0.14)

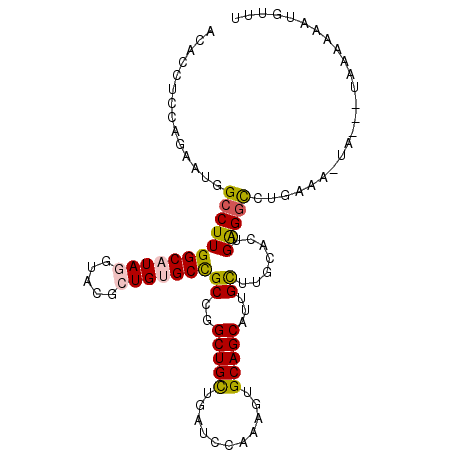
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:57 2006