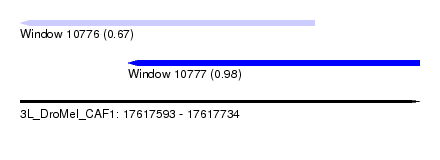
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,617,593 – 17,617,734 |
| Length | 141 |
| Max. P | 0.976935 |

| Location | 17,617,593 – 17,617,697 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.74 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17617593 104 - 23771897 CUAGCAUCCACUUGCUUUCCGCACCUGUGGAUGGUAUUUGUCACCUUAUCAAUAC-----GGUUUCUCUGUGUUGACUGUGAUUUU-AGAUAUUCCUGCGCACGAAA-AAA ....(((((((.(((.....)))...)))))))(((((((((((....(((((((-----((.....)))))))))..)))))...-))))))..............-... ( -28.60) >DroSec_CAF1 29893 103 - 1 CUAGCAUCCACAUGCUUUCCGCACCUGUGGAUGGUAUUCGUCACCUUAUCAAUAC-----GAUUUCUCCUUGUUGACUUU-AUUUU-AGAUAUUCCUGCGCACGAAA-AAA .((.(((((((((((.....)))..)))))))).))((((((......((((((.-----(.......).))))))....-....(-((......))).).))))).-... ( -21.80) >DroEre_CAF1 31414 106 - 1 UUAGCAUCCACUAGCGUUCCGCACAUGUGGAUGGUAUUCGUCACCUUAUCAAUACUGUUUGCUUUCUCUGUGUUGAAUU----UUU-AAAUAUUUCUGUGCACGGAAAACA .((.(((((((..(((...)))....))))))).))............(((((((..............)))))))...----...-.....((((((....))))))... ( -23.94) >DroYak_CAF1 31793 107 - 1 CUAGCAUCCACUUACUUUCCGCACCUGUGGAUGGUAUUCGUCACCUUAUCAAUACUGUUAGGUUUCUCUGUGUUGAUUU----UUAGAUAUAUUUUUGGGCACGAAACAAA .((.(((((((...............))))))).))(((((..((..((((((((..............))))))))..----..............))..)))))..... ( -22.27) >consensus CUAGCAUCCACUUGCUUUCCGCACCUGUGGAUGGUAUUCGUCACCUUAUCAAUAC_____GGUUUCUCUGUGUUGACUU____UUU_AGAUAUUCCUGCGCACGAAA_AAA .((.(((((((.(((.....)))...))))))).))((((((......(((((((..............)))))))............(.........)).)))))..... (-15.99 = -15.74 + -0.25)



| Location | 17,617,631 – 17,617,734 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.20 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17617631 103 - 23771897 CACAUCUGACUCAUC---CACAGGUAACUGUAAUUACUGCCUAGCAUCCACU----UGCUUUCCGCACCUGUGGAUGGUAUUUGU-CACCUUAUCAAUAC-----GGUUUCUCUGU ......(((((((((---(((((((....((....)).((..((((......----))))....)))))))))))))).....))-))..........((-----((.....)))) ( -27.60) >DroSec_CAF1 29930 103 - 1 CACAUCUGACUCAUC---CACAGGUAACUGUAAUUACUGCCUAGCAUCCACA----UGCUUUCCGCACCUGUGGAUGGUAUUCGU-CACCUUAUCAAUAC-----GAUUUCUCCUU ......(((((((((---(((((((....((....)).((..(((((....)----))))....)))))))))))))).....))-))............-----........... ( -26.20) >DroEre_CAF1 31449 108 - 1 CACUUCUGACUCAUC---CGCAGGUAACUGUAAUUACUACUUAGCAUCCACU----AGCGUUCCGCACAUGUGGAUGGUAUUCGU-CACCUUAUCAAUACUGUUUGCUUUCUCUGU ......((((.....---.((((....))))..........((.(((((((.----.(((...)))....))))))).))...))-))............................ ( -21.30) >DroYak_CAF1 31829 108 - 1 CACAUCUGACUCAUC---CGCAGGUAACUGUAAUUACUGCCUAGCAUCCACU----UACUUUCCGCACCUGUGGAUGGUAUUCGU-CACCUUAUCAAUACUGUUAGGUUUCUCUGU ...(((((((.((((---(((((((....((((....(((...))).....)----))).......)))))))))))(((((.((-......)).))))).)))))))........ ( -26.70) >DroAna_CAF1 29129 88 - 1 --CAUCUGAGUCACCCUACGCAGGUAACUGUAGUUACA--CUGGCA-----A----UACGGUCCACACUUGUGGAUCGCAUUCGG-CACCUUAUCUU------------UCUGC-- --...((((((........(((((((((....))))).--)).)).-----.----..((((((((....)))))))).))))))-...........------------.....-- ( -24.10) >DroPer_CAF1 32396 106 - 1 -ACAUCUGACUCAUC---CACAGGUAACUGUAAUUAC--CCUAGCAUCCACACAUAUGAAAUCCACACUUGUGGAAGCCAUUCGCCCACUUUAUCAAUACCGUUCA----AAGCUU -.....(((......---.((((....))))......--....((.((((((....((.....))....)))))).))...............)))..........----...... ( -14.00) >consensus CACAUCUGACUCAUC___CACAGGUAACUGUAAUUACUGCCUAGCAUCCACA____UGCUUUCCGCACCUGUGGAUGGUAUUCGU_CACCUUAUCAAUAC_____G_UUUCUCCGU ...................((((....)))).........................((((.(((((....))))).)))).................................... (-12.23 = -12.20 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:55 2006