
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
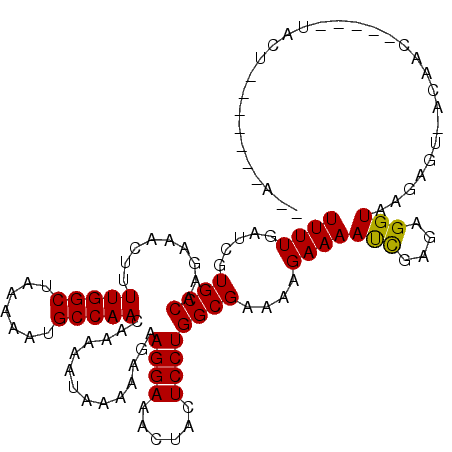
| Location | 17,606,265 – 17,606,411 |
| Length | 146 |
| Max. P | 0.999894 |

| Location | 17,606,265 – 17,606,371 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.47 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.70 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17606265 106 - 23771897 UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACCAAAAUAAAAAGAAGGAACCCACUCCUGGCGAAAAGAAAAUCGAGAGGUAAGACUUACAAC-----AACU-------U-- ((((((((.((((.........(((((.......)))))..............((((......))))))))....))...))))))...............-----....-------.-- ( -18.30) >DroVir_CAF1 38822 114 - 1 UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACACUAAUAAAAAGAAGGAAAUUACUCCUGGCGAAAAGAAAAUCGAGAGGUAAGAGU-ACAAC-----UACUAUAAAAAAUC ((((((((.((((.........(((((.......)))))..............((((......))))))))....))...))))))......(((-(....-----)))).......... ( -19.10) >DroGri_CAF1 25347 111 - 1 UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACACUAAUAAAAAGAAGGAAAUUACUCCUGGCGAAAAGAAAAUUGAGAGGUAAGAGU-ACAAC-----CACUACUAC-UA-- (((..(((.((((.........(((((.......)))))..............((((......))))))))....))...)..)))((((..(((-.....-----.)))..)))-).-- ( -19.80) >DroWil_CAF1 33235 111 - 1 UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACCAAAAUAAAAAGAAGGAACCUACUCCUGGCGAAAAGAAAAUCGAGAGGUAAGAGUAAUUACAAAAAAACU-------A-- ((((((((.((((.........(((((.......)))))..............((((......))))))))....))...)))))).((((......)))).........-------.-- ( -20.10) >DroMoj_CAF1 24330 113 - 1 UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACACUAAUAAAAAGAAGGAAAUUACUCCUGGCGAAAAGAAAAUUGAGAGGUAAGAGU-ACAAC-----UACUAUAAA-AAUC (((..(((.((((.........(((((.......)))))..............((((......))))))))....))...)..)))......(((-(....-----)))).....-.... ( -16.90) >DroAna_CAF1 20218 103 - 1 UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACCAAAAUAAAAAGAAGGAAACUACUCCUGGCGAAAAGAAAACCGAGAGGUAAGACUUA---C-----UACU-------U-- ((((.....(((((.((.....(((((.......)))))..............((....))..)).)))))....))))(((....)))........---.-----....-------.-- ( -21.60) >consensus UUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACAAAAAUAAAAAGAAGGAAACUACUCCUGGCGAAAAGAAAAUCGAGAGGUAAGAGU_ACAAC_____UACU_______A__ ((((.....((((.........(((((.......)))))..............((((......))))))))....))))(((....)))............................... (-18.06 = -17.70 + -0.36)


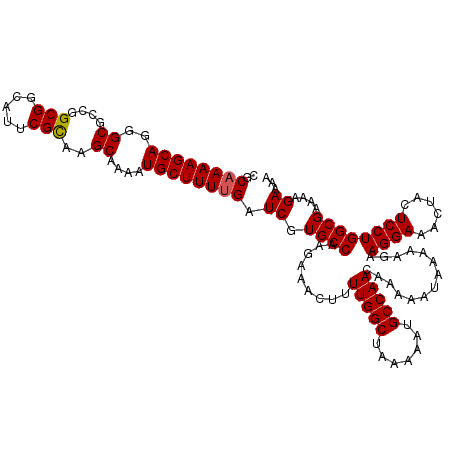
| Location | 17,606,291 – 17,606,411 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -30.47 |
| Energy contribution | -30.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17606291 120 - 23771897 CGCCAAAGCAGGGCGGCGGCGGUAUUCGCAAGCAAAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACCAAAAUAAAAAGAAGGAACCCACUCCUGGCGAAAAGAAAA ((((......((..((((((((...(((.((((.....)))).)))))))(((((((...)))))))......))))..))............((((......))))))))......... ( -34.00) >DroVir_CAF1 38856 120 - 1 CACAAAAGCAGGGCGCCGGCGGCAUUCGUAAGCAGAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACACUAAUAAAAAGAAGGAAAUUACUCCUGGCGAAAAGAAAA .............((((((((((((((.......))))))).........(((((((...))))))).......)))................((((......))))))))......... ( -31.50) >DroPse_CAF1 33232 120 - 1 CGCAAAAGCAGGGCGGCGGCGGUAUUCGCAAGCAAAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCCAAAAAUGCCAACCAAAAUAAAAAGAAGGAACCUACUCCUGGCGAAAAGAAAA ..((((((((..((....(((.....)))..))....)))))))).((.((((.........(((((.......)))))..............((((......))))))))....))... ( -30.80) >DroGri_CAF1 25378 120 - 1 CUCAAAAGCAGGGCGCCGGCGGCAUUCGUAAGCAGAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACACUAAUAAAAAGAAGGAAAUUACUCCUGGCGAAAAGAAAA .(((((((((....((..(((.....)))..))....)))))))))((.((((.........(((((.......)))))..............((((......))))))))....))... ( -31.90) >DroMoj_CAF1 24363 120 - 1 CACAAAAGCAGGGCGCCGGCGGCAUUCGUAAGCAGAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACACUAAUAAAAAGAAGGAAAUUACUCCUGGCGAAAAGAAAA .............((((((((((((((.......))))))).........(((((((...))))))).......)))................((((......))))))))......... ( -31.50) >DroPer_CAF1 21742 120 - 1 CGCAAAAGCAGGGCGGCGGCGGUAUUCGCAAGCAAAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCCAAAAAUGCCAACCAAAAUAAAAAGAAGGAACCUACUCCUGGCGAAAAGAAAA ..((((((((..((....(((.....)))..))....)))))))).((.((((.........(((((.......)))))..............((((......))))))))....))... ( -30.80) >consensus CGCAAAAGCAGGGCGCCGGCGGCAUUCGCAAGCAAAAUGCUUUUGAUCGUGCCAAGAAACUUUUGGCUAAAAAUGCCAACAAAAAUAAAAAGAAGGAAACUACUCCUGGCGAAAAGAAAA ..((((((((..((....(((.....)))..))....)))))))).((.((((.........(((((.......)))))..............((((......))))))))....))... (-30.47 = -30.38 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:50 2006