
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,600,204 – 17,600,326 |
| Length | 122 |
| Max. P | 0.968703 |

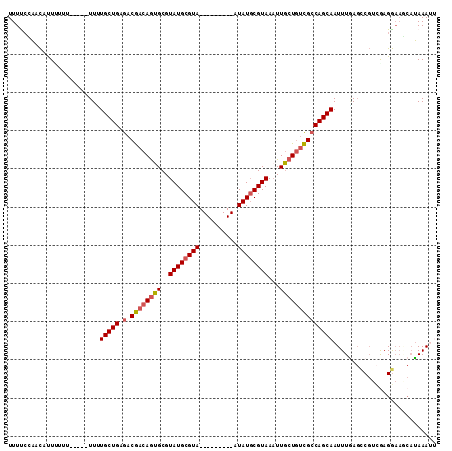
| Location | 17,600,204 – 17,600,302 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.53 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17600204 98 + 23771897 UUUCCCAACAUUUUUU-----UUUUGCUGAGACGACAGUGCGUAUGCGUA---------AUAUGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCAAGGAAGCAUAAAUU (((((...........-----..((((((.(.((((((((..(((((((.---------..)))))))...))))))))))))))).((((....)))))))))........ ( -29.00) >DroSec_CAF1 12554 100 + 1 UUUUCCAACAUUUUUUUU-UUUUUUGCUGAGACGACAGUGCGUAUGCGUA---------AUAUGCGUAAAUUGCUGUCGCCAGCAAUUUAAGACGUCGAGGAAGCAUAAA-- .(((((............-....((((((.(.((((((((..(((((((.---------..)))))))...))))))))))))))).....(....)..)))))......-- ( -28.00) >DroSim_CAF1 12622 101 + 1 UUUUCCAACAUUUUUUUUCUUUUUUGCUGAGACGACAGUGCGUAUGCGUA---------AUAUGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCGCGGAAGCAUAAA-- ..................(((..((((((.(.((((((((..(((((((.---------..)))))))...)))))))))))))))...))).....((....)).....-- ( -31.20) >DroEre_CAF1 12205 94 + 1 UUUCGUAACUUU---------UUCUGCUGAGACGACAGUGCGUAUGCGUA---------AUAUGCGUAAUUUGCUGUCGCCAGCAAUUUGAGCCGUGGAGGAAACAUAAAUU .((((...(((.---------...(((((.(.((((((((..(((((((.---------..)))))))...))))))))))))))....)))...))))(....)....... ( -27.80) >DroYak_CAF1 12607 103 + 1 UUUUCGAACAUU---------UUGUGCUGAGACGACAGUGCGUAUGCGUAAUACGUGUAAUAUGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCGAGGAAUCAUAAAUU (((((((...((---------...(((((.(.((((((((..((((((((.((....)).))))))))...))))))))))))))....))....))))))).......... ( -33.70) >DroAna_CAF1 14172 96 + 1 UUUUCUU-UACUCCUUAU-UUUUAUGCUGAGACAACAAGGCGUAUGCGUA---------AUAUCCGUAAAUUUCUAGUGACAGCAAUUUUAUUCCCUGAGA-----GAAAUU .((((((-(.........-.....(((((.....((.(((.((.((((..---------.....)))).)).))).))..))))).............)))-----)))).. ( -14.23) >consensus UUUUCCAACAUUUUUU_____UUUUGCUGAGACGACAGUGCGUAUGCGUA_________AUAUGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCGAGGAAGCAUAAAUU ........................(((((.(.((((((((..((((((((..........))))))))...))))))))))))))........................... (-21.25 = -21.53 + 0.28)


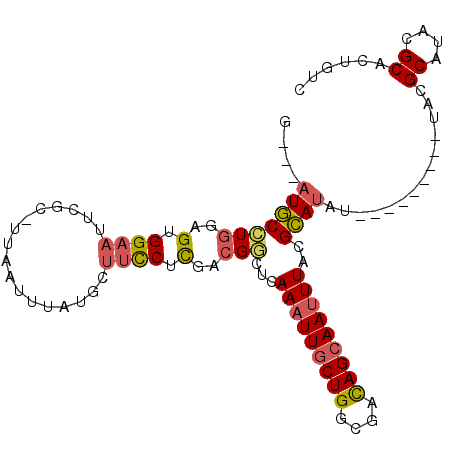
| Location | 17,600,204 – 17,600,302 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17600204 98 - 23771897 AAUUUAUGCUUCCUUGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAU---------UACGCAUACGCACUGUCGUCUCAGCAAAA-----AAAAAAUGUUGGGAAA .........((((..((((((..(((((((((....)))))))))........---------...((....))...)))))).(((((...-----......))))))))). ( -28.60) >DroSec_CAF1 12554 100 - 1 --UUUAUGCUUCCUCGACGUCUUAAAUUGCUGGCGACAGCAAUUUACGCAUAU---------UACGCAUACGCACUGUCGUCUCAGCAAAAAA-AAAAAAAAUGUUGGAAAA --....((((....((((((..((((((((((....)))))))))).))....---------...((....))...))))....)))).....-.................. ( -25.20) >DroSim_CAF1 12622 101 - 1 --UUUAUGCUUCCGCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAU---------UACGCAUACGCACUGUCGUCUCAGCAAAAAAGAAAAAAAAUGUUGGAAAA --....((((...(((((((...(((((((((....)))))))))........---------...((....)).)))))))...))))........................ ( -27.70) >DroEre_CAF1 12205 94 - 1 AAUUUAUGUUUCCUCCACGGCUCAAAUUGCUGGCGACAGCAAAUUACGCAUAU---------UACGCAUACGCACUGUCGUCUCAGCAGAA---------AAAGUUACGAAA ..................((((....((((((((((((((.......))....---------...((....))..))))))..))))))..---------..))))...... ( -18.50) >DroYak_CAF1 12607 103 - 1 AAUUUAUGAUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAUUACACGUAUUACGCAUACGCACUGUCGUCUCAGCACAA---------AAUGUUCGAAAA ...............((((((..(((((((((....)))))))))............(((((.....)))))....))))))((((((...---------..)))).))... ( -24.60) >DroAna_CAF1 14172 96 - 1 AAUUUC-----UCUCAGGGAAUAAAAUUGCUGUCACUAGAAAUUUACGGAUAU---------UACGCAUACGCCUUGUUGUCUCAGCAUAAAA-AUAAGGAGUA-AAGAAAA ((((((-----(..(((.((......)).))).....))))))).........---------......(((.(((((((.............)-)))))).)))-....... ( -14.02) >consensus AAUUUAUGCUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAU_________UACGCAUACGCACUGUCGUCUCAGCAAAA_____AAAAAAUGUUGGAAAA ......((((....((((((...(((((((((....)))))))))....................((....)).))))))....))))........................ (-17.58 = -18.30 + 0.72)



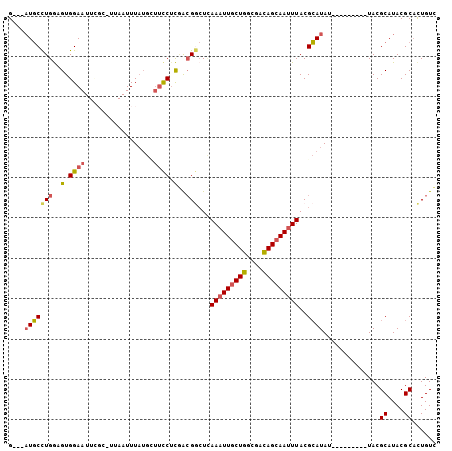
| Location | 17,600,232 – 17,600,326 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17600232 94 - 23771897 G---AUGCCUGAAGUGGAAUUCGCGUUAAUUUAUGCUUCCUUGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAU---------UACGCAUACGCACUGUC .---.(((..(((......)))((((.....(((((..((.....))...(((((((((....)))))))))..))))).---------.))))....)))..... ( -26.00) >DroSec_CAF1 12586 85 - 1 GG--AUGCCUGGAGUGGAA----------UUUAUGCUUCCUCGACGUCUUAAAUUGCUGGCGACAGCAAUUUACGCAUAU---------UACGCAUACGCACUGUC ((--(.((.((((......----------)))).)).)))..(((((..((((((((((....)))))))))).))....---------...((....))...))) ( -23.80) >DroSim_CAF1 12655 85 - 1 GG--AUGCCUGGAGUGGAA----------UUUAUGCUUCCGCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAU---------UACGCAUACGCACUGUC ..--(((((((..((((((----------.......))))))..)))...(((((((((....)))))))))..))))..---------...((....))...... ( -28.10) >DroEre_CAF1 12229 94 - 1 G---AUGCCUGGCGUGGAAUUCGCAUUAAUUUAUGUUUCCUCCACGGCUCAAAUUGCUGGCGACAGCAAAUUACGCAUAU---------UACGCAUACGCACUGUC .---((((..(.((((((....((((......))))....)))))).).....((((((....)))))).....))))..---------...((....))...... ( -27.60) >DroYak_CAF1 12631 103 - 1 G---AUGCCUGGUGUGGAAUUCGCAUUAAUUUAUGAUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAUUACACGUAUUACGCAUACGCACUGUC (---(.(((...((.(((((...(((......)))))))).))..)))))(((((((((....)))))))))............(((((.....)))))....... ( -26.20) >DroAna_CAF1 14203 92 - 1 GAACCUCCUUGGUGUGGAAUUUGCCUUAAUUUC-----UCUCAGGGAAUAAAAUUGCUGUCACUAGAAAUUUACGGAUAU---------UACGCAUACGCCUUGUU .......(..((((((.(((...((..((((((-----(..(((.((......)).))).....)))))))...))..))---------).....))))))..).. ( -16.30) >consensus G___AUGCCUGGAGUGGAAUUCGC_UUAAUUUAUGCUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAU_________UACGCAUACGCACUGUC ....(((((((..(.((((.................)))).)..)))...(((((((((....)))))))))..))))..............((....))...... (-16.04 = -16.30 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:47 2006