| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,577,492 – 17,577,590 |
| Length | 98 |
| Max. P | 0.982963 |

| Location | 17,577,492 – 17,577,590 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.64 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
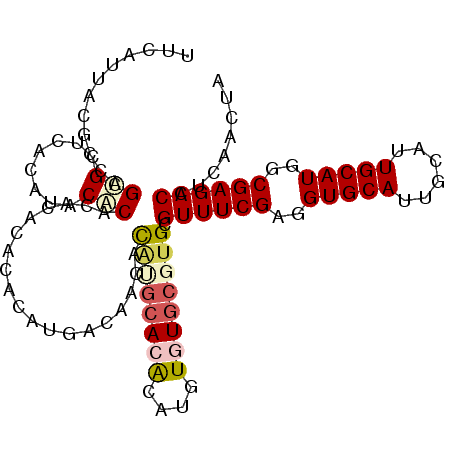
>3L_DroMel_CAF1 17577492 98 + 23771897 UUCAUUACGCCAGCGCUCACAUACGCAC----ACACAUGCAAACAC----ACAUGUGUGUGCGUGCGUUUCGAGGUGCAUUGCAUUGCAUGGCGAGACCUCAACUA .......((((....(((((.(((((((----(((((((.......----.)))))))))))))).))...)))(((((......)))))))))............ ( -39.20) >DroSec_CAF1 13904 104 + 1 UUCAUUACUCUAGCGCUCACAUACACAC--ACACACAUGACAACACAUGCACGCAUGUAUGCGUGCGUUUCGAGGUGCAUUGUAUUGCAUGGCGAGACCUCAACUA ............................--.......(((........(((((((....)))))))((((((..(((((......)))))..)))))).))).... ( -27.20) >DroSim_CAF1 16206 106 + 1 UUCAUUACGCCAGCGCUCACAUACACACACACACACAUGACAACACAUGCACGCAUGUAUGCGUGCGUUUCGAGGUGCAUUGCAUUGCAUGGCGAGACCUCAACUA .......((((((((((.....((............(((......)))(((((((....))))))))).....)))))..(((...))))))))............ ( -28.00) >DroEre_CAF1 22672 102 + 1 UCCAUUACGGCAGUGUGUACAGGCACAGACAGGCACACGCC----UGGGCACACAAGUGUGCGUGCGUUUCGAGGUGCAUUGCAUUGCAUGGCGAGACCUCAACUA ...........(((........((((...(((((....)))----)).(((((....)))))))))((((((..(((((......)))))..))))))....))). ( -38.10) >DroYak_CAF1 23562 102 + 1 UCCAUUACGCCAGUGUGCUCACACACACACACCAACACAUA----CACGCACAUAAGUGUGCGUGCGUUUCGAGGUGCAUUGCAUUGCAUGGCGAGACCUCAACUA ............(((((......))))).............----((((((((....)))))))).((((((..(((((......)))))..))))))........ ( -33.20) >consensus UUCAUUACGCCAGCGCUCACAUACACACACACACACAUGACAACACAUGCACACAUGUGUGCGUGCGUUUCGAGGUGCAUUGCAUUGCAUGGCGAGACCUCAACUA ............(((........)))...................((((((((....)))))))).((((((..(((((......)))))..))))))........ (-20.60 = -21.64 + 1.04)


| Location | 17,577,492 – 17,577,590 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.26 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17577492 98 - 23771897 UAGUUGAGGUCUCGCCAUGCAAUGCAAUGCACCUCGAAACGCACGCACACACAUGU----GUGUUUGCAUGUGU----GUGCGUAUGUGAGCGCUGGCGUAAUGAA ...(((((((...((..(((...)))..))))))))).((((((((((((((((((----......))))))))----))))))..((....))..))))...... ( -39.40) >DroSec_CAF1 13904 104 - 1 UAGUUGAGGUCUCGCCAUGCAAUACAAUGCACCUCGAAACGCACGCAUACAUGCGUGCAUGUGUUGUCAUGUGUGU--GUGUGUAUGUGAGCGCUAGAGUAAUGAA ...(((((((........(((......))))))))))..(((.((((((((((((..((((((....))))))..)--))))))))))).)))............. ( -38.50) >DroSim_CAF1 16206 106 - 1 UAGUUGAGGUCUCGCCAUGCAAUGCAAUGCACCUCGAAACGCACGCAUACAUGCGUGCAUGUGUUGUCAUGUGUGUGUGUGUGUAUGUGAGCGCUGGCGUAAUGAA ........((..(((((.((..(((...))).((((....(((((((((((((((((((.....)).)))))))))))))))))...)))).)))))))..))... ( -40.50) >DroEre_CAF1 22672 102 - 1 UAGUUGAGGUCUCGCCAUGCAAUGCAAUGCACCUCGAAACGCACGCACACUUGUGUGCCCA----GGCGUGUGCCUGUCUGUGCCUGUACACACUGCCGUAAUGGA ...(((((((...((..(((...)))..)))))))))...(((((((....)))))))(((----(((((((((..((....))..))))))...)))....))). ( -31.10) >DroYak_CAF1 23562 102 - 1 UAGUUGAGGUCUCGCCAUGCAAUGCAAUGCACCUCGAAACGCACGCACACUUAUGUGCGUG----UAUGUGUUGGUGUGUGUGUGUGAGCACACUGGCGUAAUGGA ...(((((((...((..(((...)))..))))))))).(((((((((((....))))))))----)....((..((((((........))))))..))))...... ( -38.10) >consensus UAGUUGAGGUCUCGCCAUGCAAUGCAAUGCACCUCGAAACGCACGCACACAUGUGUGCAUGUGUUGGCAUGUGUGUGUGUGUGUAUGUGAGCGCUGGCGUAAUGAA ((((((((((...((..(((...)))..))))))))....(((((((((((((((((..........)))))))))))))))))........)))).......... (-22.50 = -23.26 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:39 2006