
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,551,250 – 17,551,344 |
| Length | 94 |
| Max. P | 0.659459 |

| Location | 17,551,250 – 17,551,344 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -11.38 |
| Energy contribution | -12.55 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
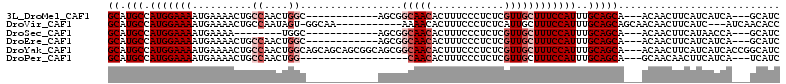
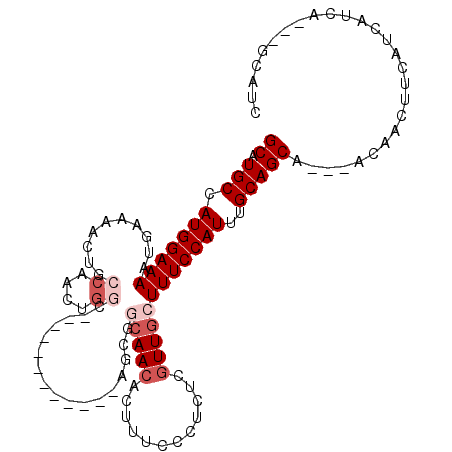
>3L_DroMel_CAF1 17551250 94 - 23771897 GCAUGCCAUGGAAAAUGAAAACUGCCAACUGGC------------AGCGGCAACACUUUCCCUCUCGUUGCUUUCCAUUUGCAGCA---ACAACUUCAUCAUCA---GCAUC ((.(((.(((((((.......(((((....)))------------))..(((((............))))))))))))..))))).---...............---..... ( -25.00) >DroVir_CAF1 35429 97 - 1 GCAUGCCAUGGAAAAUGAAAACUGCCAAUAGU-GGCAA-----------AAAACACUUUCCCUCUCAUUGCUUUCCAUUUGCAGCAGCAACAACUUCAUC---AUCAACACC ((.(((...(((((.((.....(((((....)-)))).-----------....)).))))).......(((.........))))))))............---......... ( -18.00) >DroSec_CAF1 36827 86 - 1 GCAUGCCAUGGAAAAUGAAAA--------UGGC------------AGCGGCAACACUUUCCCUCUCGUUGCUUUCCAUUUGCAGCA---ACAACUUCAUAACCA---GCAUC ((.(((.(((((((((....)--------).((------------(((((..............))))))))))))))..))))).---...............---..... ( -18.34) >DroEre_CAF1 36761 94 - 1 GCAUGCCAUGGAAAAUGAAAACUGCCAACUGGC------------AGCGGCAACACUUUCCCUCUCGUUGCUUUCCAUUUGCAGCA---ACAACUUCAUCAUCA---GCAUC ((.(((.(((((((.......(((((....)))------------))..(((((............))))))))))))..))))).---...............---..... ( -25.00) >DroYak_CAF1 40440 109 - 1 GCAUGCCAUGGAAAAUGAAAACUGCCAACUGGCAGCAGCAGCGGCAGCGGCAACACUUUCCCUCUCGUUGCUUUCCAUUUGCAGCA---ACAACUUCAUCAUCACCGGCAUC ..(((((..(((((.((....(((((....)))))..((.((....)).))..)).))))).....((((((..........))))---))...............))))). ( -32.10) >DroPer_CAF1 45511 88 - 1 GCAUGCCAUGGAAAAUGAAAACUGCCAACUGG------------------CAACACUUUCCCUCUCGUUGCUUUCCAUUUGCAGCA---GCAACAACUUCAUCA---UCAUC ...(((...(((((.((.....((((....))------------------)).)).))))).....(((((.........))))).---)))............---..... ( -17.70) >consensus GCAUGCCAUGGAAAAUGAAAACUGCCAACUGGC____________AGCGGCAACACUUUCCCUCUCGUUGCUUUCCAUUUGCAGCA___ACAACUUCAUCAUCA___GCAUC ((.(((.(((((((..........((....)).................(((((............))))))))))))..)))))........................... (-11.38 = -12.55 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:31 2006