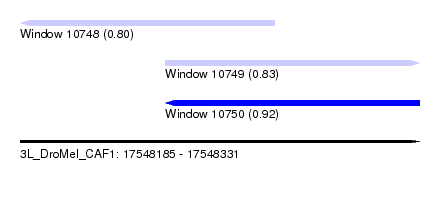
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,548,185 – 17,548,331 |
| Length | 146 |
| Max. P | 0.915612 |

| Location | 17,548,185 – 17,548,278 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -14.01 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17548185 93 - 23771897 CUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUGCAUUUGC-----------GGCUUGGUCAUCCAACCUGGUGG-----CCAAAGCCAAAUCAUGGGUU ((((((....))))))((((.((((.((((..((........))..))))-----------((((((((((((......)))))-----)).))))).)))).)))).. ( -34.80) >DroSec_CAF1 34098 92 - 1 CUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUGCAGUGGC-----------GGCUUGGUCAUCCAACCUGGUGG-----CCA-AGCCAAAUCAUGGGUU ((((((....))))))((((.((((.((....))..(((((....)))))-----------((((((((((((......)))))-----)))-)))).)))).)))).. ( -41.70) >DroSim_CAF1 36583 93 - 1 CUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUGCAGUUGC-----------GGCUUGGUCAUCCAACCUGGUGG-----CCAAAGCCAAAUCAUGGGUU ((((((....))))))((((.((((.(((((.((........)).)))))-----------((((((((((((......)))))-----)).))))).)))).)))).. ( -36.40) >DroEre_CAF1 33890 97 - 1 CUGCAUACAAAUGCGGCUCGAGAUUCGCGAUCGCUGGCCACUGCAGUGGC-----------GGCUUGGUCAUCCAACCUGGUGGCCUGGCCAGAGCCAAAUCAUGG-UU ..((((....))))(((((.......((....))..(((((....)))))-----------((((.(((((((......))))))).)))).))))).........-.. ( -40.80) >DroYak_CAF1 37041 97 - 1 CUGCAUACAAAUGCG-------AUUCGCGAUCGCUGACUAUUGCAGUGGCUUGUGGCUUGUGGCUUGGUCAUCCAACCUGGUGG-----CCAGCUCCAAAUCAUGGCUU (((((.......(((-------(((...)))))).......))))).((((.(((..(((..(((.(((((((......)))))-----)))))..)))..))))))). ( -36.24) >DroPer_CAF1 42172 79 - 1 CUGCAUACAAAUGCGACUGCA-----GCGAUUGCCGGCCAUCGCUGUAGU-----------CUCUUGGUCAUCCAACCUGG--------------CCAGAUCAUGGGGG ((.(((........(((((((-----(((((.(....).)))))))))))-----------)..(((((((.......)))--------------))))...))).)). ( -31.30) >consensus CUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUGCAGUGGC___________GGCUUGGUCAUCCAACCUGGUGG_____CCA_AGCCAAAUCAUGGGUU ((((((....))))))..........((....)).(((((..((..................)).)))))..(((...((((((.....)))...))).....)))... (-14.01 = -14.10 + 0.10)

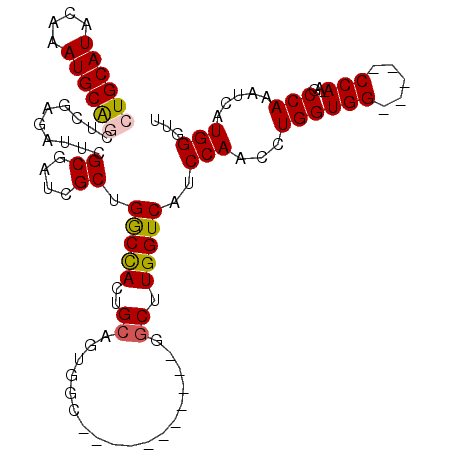
| Location | 17,548,238 – 17,548,331 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.40 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17548238 93 + 23771897 UGGCCAGCGAUCGCGAAUCUCGAGCUGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGACCAUUCAGCCAUUUGGCCAUUCAUAAUGAAAGUGA ((((((((((...(((((((((((((((((....))))))).....))))).))))).....)).))....)))))).((((.......)))) ( -32.20) >DroPse_CAF1 37893 72 + 1 UGGCCGGCAAUCGC-----UGCAGUCGCAUUUGUAUGCAGCGCCAUGCGA--------CCG--------GCCGUCCAUUCAUAAUGAAAGUGA .((((((...((((-----.((.((.((((....)))).))))...))))--------)))--------)))......((((.......)))) ( -25.30) >DroSec_CAF1 34150 85 + 1 UGGCCAGCGAUCGCGAAUCUCGAGCUGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGACCG--------UUUGGCCAUUCAUAAUGAAAGUGA (((((((((....(((((((((((((((((....))))))).....))))).)))))..))--------.))))))).((((.......)))) ( -33.90) >DroSim_CAF1 36636 85 + 1 UGGCCAGCGAUCGCGAAUCUCGAGCUGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGACCG--------UUUGGCCAUUCAUAAUGAAAGUGA (((((((((....(((((((((((((((((....))))))).....))))).)))))..))--------.))))))).((((.......)))) ( -33.90) >DroEre_CAF1 33947 85 + 1 UGGCCAGCGAUCGCGAAUCUCGAGCCGCAUUUGUAUGCAGCGCCAUUCGAGCAUUCGGCCA--------UUUGGCCAUUCAUAAUGAAAGUGA ((((((((....))(((((((((((.((((....)))).)).....))))).)))).....--------..)))))).((((.......)))) ( -26.70) >DroPer_CAF1 42216 72 + 1 UGGCCGGCAAUCGC-----UGCAGUCGCAUUUGUAUGCAGCGCCAUGCGA--------CCG--------GCCGUCCAUUCAUAAUGAAAGUGA .((((((...((((-----.((.((.((((....)))).))))...))))--------)))--------)))......((((.......)))) ( -25.30) >consensus UGGCCAGCGAUCGCGAAUCUCGAGCCGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGACCG________UUUGGCCAUUCAUAAUGAAAGUGA ((((((((....))(((((((((((.((((....)))).)).....))))).))))...............)))))).((((.......)))) (-16.32 = -17.40 + 1.08)


| Location | 17,548,238 – 17,548,331 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -17.96 |
| Energy contribution | -19.35 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17548238 93 - 23771897 UCACUUUCAUUAUGAAUGGCCAAAUGGCUGAAUGGUCGAAUGCUCGAGUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCA ................((((((...((((....))))((((.(((((.....(((((((....))))))))))))))))((....)))))))) ( -32.50) >DroPse_CAF1 37893 72 - 1 UCACUUUCAUUAUGAAUGGACGGC--------CGG--------UCGCAUGGCGCUGCAUACAAAUGCGACUGCA-----GCGAUUGCCGGCCA .....(((((.....))))).(((--------(((--------(.......(((((((..(......)..))))-----)))...))))))). ( -26.20) >DroSec_CAF1 34150 85 - 1 UCACUUUCAUUAUGAAUGGCCAAA--------CGGUCGAAUGCUCGAGUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCA ................((((((..--------((((((....(((((.....(((((((....)))))))))))).....)))))).)))))) ( -34.20) >DroSim_CAF1 36636 85 - 1 UCACUUUCAUUAUGAAUGGCCAAA--------CGGUCGAAUGCUCGAGUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCA ................((((((..--------((((((....(((((.....(((((((....)))))))))))).....)))))).)))))) ( -34.20) >DroEre_CAF1 33947 85 - 1 UCACUUUCAUUAUGAAUGGCCAAA--------UGGCCGAAUGCUCGAAUGGCGCUGCAUACAAAUGCGGCUCGAGAUUCGCGAUCGCUGGCCA ................((((((.(--------(.((.((((.(((((.....(((((((....)))))))))))))))))).))...)))))) ( -30.20) >DroPer_CAF1 42216 72 - 1 UCACUUUCAUUAUGAAUGGACGGC--------CGG--------UCGCAUGGCGCUGCAUACAAAUGCGACUGCA-----GCGAUUGCCGGCCA .....(((((.....))))).(((--------(((--------(.......(((((((..(......)..))))-----)))...))))))). ( -26.20) >consensus UCACUUUCAUUAUGAAUGGCCAAA________CGGUCGAAUGCUCGAAUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCA ................((((((...............((((.(((((.....(((((((....))))))))))))))))((....)))))))) (-17.96 = -19.35 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:29 2006