
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,542,132 – 17,542,231 |
| Length | 99 |
| Max. P | 0.523744 |

| Location | 17,542,132 – 17,542,231 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.73 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
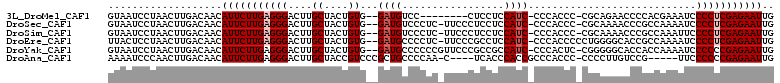
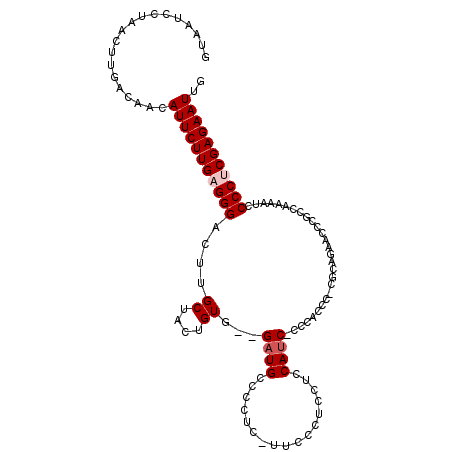
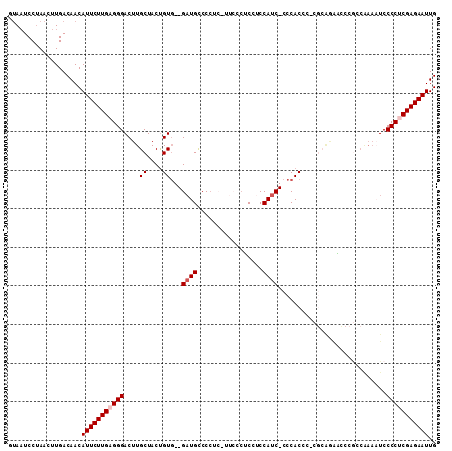
>3L_DroMel_CAF1 17542132 99 - 23771897 GUAAUCCUAACUUGACAACAUUCUUGAGGGACUUGCUACUGUG--GAUGUCC--------CUCCUCCAUC-CCCACCC-CGCAGAACCCCACGAAAUCCCCUCGAGAAUUG ...................(((((((((((..((((...((.(--((((...--------......))))-).))...-.))))........(....)))))))))))).. ( -22.80) >DroSec_CAF1 28055 106 - 1 GUAAUCCUAACUUGACAACAUUCUUGAGGGACUUGCUACUGUG--GAUGUCCCUC-UUCCCUCCUCCAUC-CCCACCC-CGCAAAACCCGCCAAAAUCCCCUCGAGAAUUG ...................(((((((((((.........((.(--((((......-..........))))-).))...-.((.......)).......))))))))))).. ( -21.69) >DroSim_CAF1 30523 106 - 1 GUAAUCCUAACUUGACAACAUUCUUGAGGGACUUGCUACUGUG--GAUGUCCCUC-UUCCCUCCUCCAUC-CCCACCC-CGCAAAACCCGCCAAAUUCCCCUCGAGAAUUG ...................(((((((((((.........((.(--((((......-..........))))-).))...-.((.......)).......))))))))))).. ( -21.69) >DroEre_CAF1 28081 107 - 1 UUACUCCUAACUUGACAACAUUCUUGAGGGACUUGCUACUGUG--GAUGCCCCUC-UUCCCGCCUCCAUC-CCCACCCCCUGGGGCACCGCCAAAAUCCCCUCGAGAAUUG ...................(((((((((((..((......(((--(.((((((..-..............-..........))))))))))...))..))))))))))).. ( -31.57) >DroYak_CAF1 30282 107 - 1 GUAAUCCUAACUUGACAACAUUCUUGAGGGACUUGCUACUGUG--GAUGCCCCCCGUUCCCGCCGCCAUC-CCCACUC-CGGGGGCACCACCAAAAUCCCCCCGAGAAUUG ...................(((((((.((((.((......(((--(.(((((((.(..............-......)-.)))))))))))..)).))))..))))))).. ( -30.25) >DroAna_CAF1 29056 100 - 1 AAAAUCCCAACUUGACAACAUUCUUGAGGGACUUGCUACCGUCCCGCUGCCCCAA-C----UCACCCACCGCCCACCC-CCCCUUGUCCG-----UUCCCCCCGAGAAUUG ...................(((((((.(((((........)))))..........-.----........((..((...-.....))..))-----.......))))))).. ( -14.80) >consensus GUAAUCCUAACUUGACAACAUUCUUGAGGGACUUGCUACUGUG__GAUGCCCCUC_UUCCCUCCUCCAUC_CCCACCC_CGCAGAACCCGCCAAAAUCCCCUCGAGAAUUG ...................(((((((((((....((....))...((((.................))))............................))))))))))).. (-13.23 = -13.73 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:24 2006