| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,960,275 – 1,960,372 |
| Length | 97 |
| Max. P | 0.969956 |

| Location | 1,960,275 – 1,960,372 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
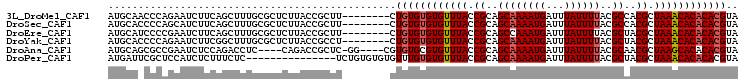
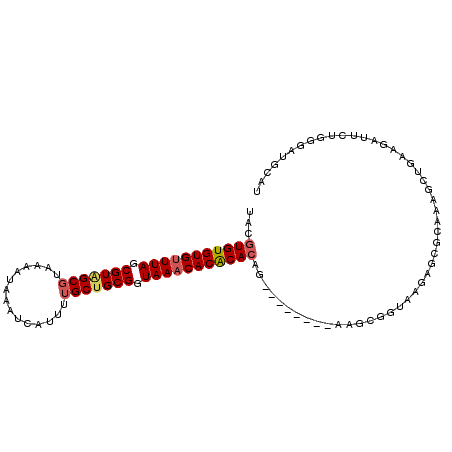
>3L_DroMel_CAF1 1960275 97 + 23771897 AUGCAACCCAGAAUCUUCAGCUUUGCGCUCUUACCGCUU--------CUGUGUGUGUUUACCGCAGCAAAAUGAUUUAUUUUACGCCACGCUAAACACACACGUA ..((((..(.((....)).)..))))((.......))..--------..(((((((((((.((..((((((((...))))))..))..)).)))))))))))... ( -21.80) >DroSec_CAF1 11479 97 + 1 AUGCACCCCAGCAUCUUCAGCUUUGCGCUCUUACCGCUU--------CUGUGUGUGUUUACCGCAGCAAAAUGAUUUAUUUUACGCCACGCUAAACACACACGUA ((((......))))..........(((.......)))..--------..(((((((((((.((..((((((((...))))))..))..)).)))))))))))... ( -23.00) >DroEre_CAF1 11276 97 + 1 AUGCAUCCCCGAAUCUUCAGCUUUGCGCUCUUACCGCUU--------CUGUGUGUGUUUACCGCAGCCAAAUGAUUUAUUUUACGCUACGCUAAACACACACGUA .(((....................(((.......)))..--------..(((((((((((.((.(((.(((........)))..))).)).)))))))))))))) ( -22.40) >DroYak_CAF1 11555 97 + 1 AUGCACCCCAGAAUCUUCGGCUUUGCGCUCUUACCGCCU--------CUGUGUGUGUUUACCGCAGCAAAAUGAUUUAUUUUACGCUACGCUAAACACACACGUA .(((..............(((..............))).--------..(((((((((((.((.(((((((((...))))))..))).)).)))))))))))))) ( -24.34) >DroAna_CAF1 9371 96 + 1 AUGCAGCGCCGAAUCUCCAGACCUC----CAGACCGCUC-GG----CGUGUGCGUGUUUACCGCAGCAAAAUGAUUUAUUUUACGCAACGCUAAGCACACACGUA .(((.(((((((...((..(....)----..))....))-))----)))(((.(((((((.((..((((((((...))))))..))..)).))))))).)))))) ( -26.20) >DroPer_CAF1 12749 90 + 1 AUGAUUCGCUCCAUCUCUUUCUC---------------UCUGUGUGUGUUUGUGUGUUUACCGCAGCAAAAUGAUUUAUUUUACGCUACGCUAAACACACACGUA .......................---------------...(((((((((((.((((.....((.(.((((((...)))))).))).)))))))))))))))... ( -19.50) >consensus AUGCACCCCAGAAUCUUCAGCUUUGCGCUCUUACCGCUU________CUGUGUGUGUUUACCGCAGCAAAAUGAUUUAUUUUACGCUACGCUAAACACACACGUA ................................................((((((((((((.((..((((((((...))))))..))..)).)))))))))))).. (-16.49 = -16.85 + 0.36)
| Location | 1,960,275 – 1,960,372 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -33.49 |
| Consensus MFE | -25.07 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
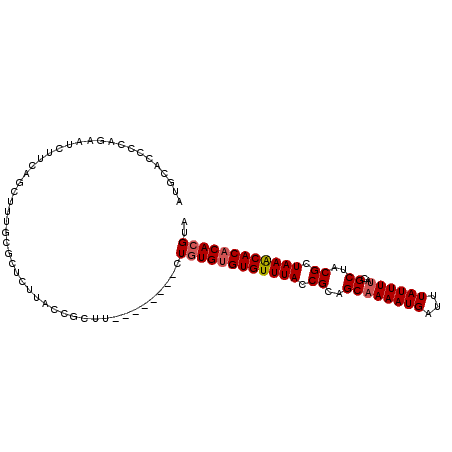
>3L_DroMel_CAF1 1960275 97 - 23771897 UACGUGUGUGUUUAGCGUGGCGUAAAAUAAAUCAUUUUGCUGCGGUAAACACACACAG--------AAGCGGUAAGAGCGCAAAGCUGAAGAUUCUGGGUUGCAU ...(((((((((((.((..(((...............)))..)).)))))))))))..--------..((.......))((((..(.((....)).)..)))).. ( -33.16) >DroSec_CAF1 11479 97 - 1 UACGUGUGUGUUUAGCGUGGCGUAAAAUAAAUCAUUUUGCUGCGGUAAACACACACAG--------AAGCGGUAAGAGCGCAAAGCUGAAGAUGCUGGGGUGCAU ...(((((((((((.((..(((...............)))..)).)))))))))))..--------...........((((..(((.......)))...)))).. ( -36.96) >DroEre_CAF1 11276 97 - 1 UACGUGUGUGUUUAGCGUAGCGUAAAAUAAAUCAUUUGGCUGCGGUAAACACACACAG--------AAGCGGUAAGAGCGCAAAGCUGAAGAUUCGGGGAUGCAU ...(((((((((((.((((((.((((........)))))))))).)))))))))))..--------..((.......))(((...((((....))))...))).. ( -35.40) >DroYak_CAF1 11555 97 - 1 UACGUGUGUGUUUAGCGUAGCGUAAAAUAAAUCAUUUUGCUGCGGUAAACACACACAG--------AGGCGGUAAGAGCGCAAAGCCGAAGAUUCUGGGGUGCAU ...(((((((((((.(((((((...............))))))).)))))))))))..--------..((.......))(((...(((.......)))..))).. ( -34.46) >DroAna_CAF1 9371 96 - 1 UACGUGUGUGCUUAGCGUUGCGUAAAAUAAAUCAUUUUGCUGCGGUAAACACGCACACG----CC-GAGCGGUCUG----GAGGUCUGGAGAUUCGGCGCUGCAU ...(((((((.(((.(((...(((((((.....))))))).))).))).))))))).((----((-(((...((..----(....)..))..)))))))...... ( -35.60) >DroPer_CAF1 12749 90 - 1 UACGUGUGUGUUUAGCGUAGCGUAAAAUAAAUCAUUUUGCUGCGGUAAACACACAAACACACACAGA---------------GAGAAAGAGAUGGAGCGAAUCAU ....((((((((((.(((((((...............))))))).))))))))))............---------------........(((.......))).. ( -25.36) >consensus UACGUGUGUGUUUAGCGUAGCGUAAAAUAAAUCAUUUUGCUGCGGUAAACACACACAG________AAGCGGUAAGAGCGCAAAGCUGAAGAUUCUGGGAUGCAU ...(((((((((((.(((((((...............))))))).)))))))))))................................................. (-25.07 = -25.43 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:53 2006