
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,497,269 – 17,497,422 |
| Length | 153 |
| Max. P | 0.911088 |

| Location | 17,497,269 – 17,497,385 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -15.41 |
| Energy contribution | -17.37 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.911088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17497269 116 - 23771897 GCAAAACAAUAGAAAAGCC-CAUUCCAGACACUAAGUAAACAACGAUGAUGCCAGUUUAUUUUCUGGCGGCUUCCUAAUACUUUUCUUGGCACUUUUCU--GCU-CUUCGGAAAGUGCAA .........(((..(((((-....(((((....((((((((.............))))))))))))).))))).)))............(((((((((.--...-....))))))))).. ( -27.42) >DroSec_CAF1 4765 116 - 1 GCAAAACAAUGGAAUAGCC-CAUUCCAGACACUAAGCAAACAACAAUGAUGCCAGUUUAUUUUCUGGCGGCUACCUAAUACUUUUCUUGGCACUUUUCG--GCU-CUUUGAAAAGUGCAA ......(((((((((....-.)))))).......(((...((....)).((((((........)))))))))..............)))((((((((((--(..-..))))))))))).. ( -28.90) >DroSim_CAF1 4473 116 - 1 GCAAAACAAUGGAAUAGCC-CAUUCCAGACACUAAGCAAACAACGAUGAUGCCAGUUUAUUUUCUGGCGGCUACCAAAUACUUUUCUUGGCACUUUUCU--GCU-CUUCGGAAAGUGCAA .........(((..(((((-((((....................))))..(((((........))))))))))))).............(((((((((.--...-....))))))))).. ( -28.45) >DroEre_CAF1 10330 89 - 1 GCAAAACAAUGGAAUAGCC-CAUUCCAGACACUAAGUAAACAAGAAUGAUGCCAGUUUAUUUUCUGGCGGUUUCCAAAUACUUU------------------------CGGAA------A .........(((((..(((-(((((...((.....))......)))))..(((((........)))))))))))))........------------------------.....------. ( -18.80) >DroYak_CAF1 10072 111 - 1 GCACAACAAUGGAAUAGCC-CAUUCCAGACACUAAGUAAACAACAAUGAUGCCAGUUUAUUUUCUGGCGGCUUCAUAAUACUUUUCUUGGCACCCUUCU--G------CGAAAAGUGCAA ((((.....(((......)-))(((((((..(((((.........((((((((((........))))))...)))).........)))))......)))--)------.)))..)))).. ( -26.17) >DroAna_CAF1 9480 101 - 1 GUCGGCCAGUCGGCGAGCCACAUUCCAGACGAGGGCCAAACA------------CUUUUUUUUC----GGCU--CGACUGCUUAUC-AGGCACUUUUCGCUGCCAACUGGAAAAGUGCAA ...((((.(((((.((......)))).)))...))))...((------------(((((((...----(((.--(((.((((....-.))))....)))..)))....)))))))))... ( -29.70) >consensus GCAAAACAAUGGAAUAGCC_CAUUCCAGACACUAAGCAAACAACAAUGAUGCCAGUUUAUUUUCUGGCGGCUUCCUAAUACUUUUCUUGGCACUUUUCU__GCU_CUUCGAAAAGUGCAA ((.......((((((......))))))......................((((((........))))))))..................(((((((((...........))))))))).. (-15.41 = -17.37 + 1.96)

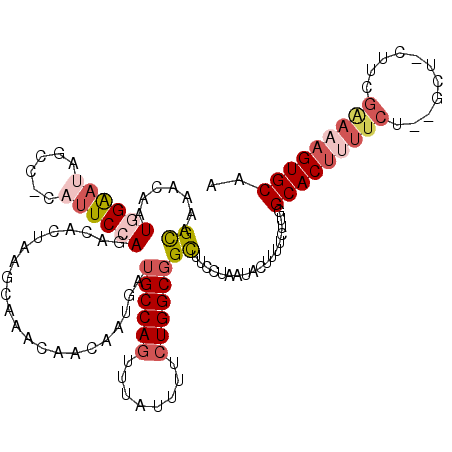
| Location | 17,497,306 – 17,497,422 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.86 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17497306 116 - 23771897 CUGAGAUGGCGAGCGAAUUAUUUGUUUAUACAAGUGAGCAAAACAAUAGAAAAGCCCAUUCCAGACACUAAGUAAACAACGAUGAUGCCAGUUUAUUUUCUGGCGGCUUCCUAAUA (((..(((((...(......(((((((((....)))))))))......)....).))))..)))................((...((((((........))))))...))...... ( -25.30) >DroSec_CAF1 4802 116 - 1 CUGAGAUGGCGAGCGAAUUAUUUGUUUAUACAAGUGAGCAAAACAAUGGAAUAGCCCAUUCCAGACACUAAGCAAACAACAAUGAUGCCAGUUUAUUUUCUGGCGGCUACCUAAUA ...((.((((..((......(((((((((....)))))))))....((((((.....))))))........))............((((((........)))))))))).)).... ( -27.90) >DroSim_CAF1 4510 116 - 1 CUGAGAUGGCGAGCGAAUUAUUUGUUUAUACAAGUGAGCAAAACAAUGGAAUAGCCCAUUCCAGACACUAAGCAAACAACGAUGAUGCCAGUUUAUUUUCUGGCGGCUACCAAAUA ....(.((((..((......(((((((((....)))))))))....((((((.....))))))........))............((((((........)))))))))).)..... ( -27.10) >DroEre_CAF1 10340 116 - 1 CUGAGAUGGCGAGCGAAUUAUUUGUUUAUACAAGUGAGCAAAACAAUGGAAUAGCCCAUUCCAGACACUAAGUAAACAAGAAUGAUGCCAGUUUAUUUUCUGGCGGUUUCCAAAUA ......(((.((((......(((((((((...((((.(.....)..((((((.....))))))..))))..))))))))).....((((((........))))))))))))).... ( -27.10) >DroYak_CAF1 10104 116 - 1 CUGAGAUGGCGAGCGAAUUAUUUGUUUAUACAAGUGAGCACAACAAUGGAAUAGCCCAUUCCAGACACUAAGUAAACAACAAUGAUGCCAGUUUAUUUUCUGGCGGCUUCAUAAUA .....(((..((((..(((((((((((((...((((.(.....)..((((((.....))))))..))))..)))))))..))))))(((((........))))).))))))).... ( -26.00) >consensus CUGAGAUGGCGAGCGAAUUAUUUGUUUAUACAAGUGAGCAAAACAAUGGAAUAGCCCAUUCCAGACACUAAGUAAACAACAAUGAUGCCAGUUUAUUUUCUGGCGGCUUCCUAAUA (((..(((((......(((.(((((((((....)))))))))..)))......).))))..))).....................((((((........))))))........... (-23.12 = -23.32 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:16 2006