| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,492,954 – 17,493,067 |
| Length | 113 |
| Max. P | 0.960258 |

| Location | 17,492,954 – 17,493,067 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -41.95 |
| Consensus MFE | -29.47 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
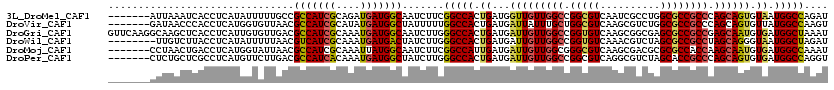
>3L_DroMel_CAF1 17492954 113 + 23771897 -------AUUAAAUCACCUCAUAUUUUUGCCGCCAUCGCAGAUGAUGGCAAUCUUCGGCCACUGAUGGUUGUUGGCCGGCGUCAAUCGCCUGGCGCCGCCCAGCAGUGUAAUGGCCAGAU -------.....(((................(((((((....))))))).......(((((.((....(((((((.((((((((......)))))))).)))))))..)).))))).))) ( -44.40) >DroVir_CAF1 371 113 + 1 -------GAUAACCCACCUCAUGGUGUUAACGCCAUCGCAUAUGAUGGCUAUUUUUGGCCACUGAUGAUUAUUUGCUGGCGUCAAGCGUCUGGCGCCGCCCAGCAGUGUUAUGGCCAAGU -------.......((((....)))).....(((((((....)))))))....((((((((....((((...(((((((.((...(((.....))).))))))))).)))))))))))). ( -41.60) >DroGri_CAF1 542 120 + 1 GUUCAAGGCAAGCUCACCUCAUUGUGUUGACGCCAUCGCAAAUGAUGGCAAUCUUGGGCCACUGAUGAUUGUUGGCCGGUGUCAAGCGGCGAGCGCCGCCGAGCAAUGUGAUGGCUAAAU ..........(((.((.(.((((((.((((((((((((....))))))).......((((((........).)))))...)))))(((((....)))))...)))))).).))))).... ( -45.30) >DroWil_CAF1 8737 112 + 1 --------UUGUCUUACCUCAUAUUUUUAACGUCAUCGCAAAUGAUGACUAUCUUGGGCCACUGAUGAUUGUUGGCCGGUGUCAAACGUCUAGCGCCGCCUAGCAGGGUAAUGGCUAGAU --------(((...((((.............(((((((....))))))).......((((((........).))))))))).)))..(((((((...((((....))))....))))))) ( -32.80) >DroMoj_CAF1 509 113 + 1 -------CCUAACUGACCUCAUGGUAUUAACGCCAUCGCAAAUUAUGGCAAUCUUCGGCCAUUGAUGAUUGUUGGCGGGCGUCAAGCGACGCGCGCCACCAAGCAAUGUGAUGGCCAAAU -------........(((....)))......(((((((((....(((((........)))))...((.(((.(((((.(((((....))))).))))).))).)).)))))))))..... ( -46.10) >DroPer_CAF1 32137 113 + 1 -------CUCUGCUCGCCUCAUGUUCUUGACGCCAUCACAAAUGAUGGCUAUCUUGGGCCACUGAUGAUUGUUGGCCGGCGUCAGGCGUCUAGCACCGCCCAGCAGUGUGAUGGCCAGGU -------........((((((......))).((((((((......(((((......)))))......((((((((.(((.(..((....))..).))).))))))))))))))))..))) ( -41.50) >consensus _______CUUAACUCACCUCAUGGUGUUAACGCCAUCGCAAAUGAUGGCAAUCUUGGGCCACUGAUGAUUGUUGGCCGGCGUCAAGCGUCUAGCGCCGCCCAGCAGUGUGAUGGCCAAAU ...............................(((((((....))))))).......(((((.((...(((((((((.(((((..........)))))))).)))))).)).))))).... (-29.47 = -29.33 + -0.14)


| Location | 17,492,954 – 17,493,067 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17492954 113 - 23771897 AUCUGGCCAUUACACUGCUGGGCGGCGCCAGGCGAUUGACGCCGGCCAACAACCAUCAGUGGCCGAAGAUUGCCAUCAUCUGCGAUGGCGGCAAAAAUAUGAGGUGAUUUAAU------- ...((((((((....((.(((.(((((.(((....))).))))).))).))......)))))))).((((..((.((((((((....)))).......))))))..))))...------- ( -41.01) >DroVir_CAF1 371 113 - 1 ACUUGGCCAUAACACUGCUGGGCGGCGCCAGACGCUUGACGCCAGCAAAUAAUCAUCAGUGGCCAAAAAUAGCCAUCAUAUGCGAUGGCGUUAACACCAUGAGGUGGGUUAUC------- ..((((((((.....((((((((((((.....))).)).).))))))...........)))))))).....((((((......)))))).....((((....)))).......------- ( -38.19) >DroGri_CAF1 542 120 - 1 AUUUAGCCAUCACAUUGCUCGGCGGCGCUCGCCGCUUGACACCGGCCAACAAUCAUCAGUGGCCCAAGAUUGCCAUCAUUUGCGAUGGCGUCAACACAAUGAGGUGAGCUUGCCUUGAAC ....(((((((.(((((...((((((....))))))((((...(((((.(........)))))).......((((((......))))))))))...))))).)))).))).......... ( -45.00) >DroWil_CAF1 8737 112 - 1 AUCUAGCCAUUACCCUGCUAGGCGGCGCUAGACGUUUGACACCGGCCAACAAUCAUCAGUGGCCCAAGAUAGUCAUCAUUUGCGAUGACGUUAAAAAUAUGAGGUAAGACAA-------- .((((((..(..((......))..).)))))).((((...((((((((.(........)))))).......((((((......)))))).............))).))))..-------- ( -30.00) >DroMoj_CAF1 509 113 - 1 AUUUGGCCAUCACAUUGCUUGGUGGCGCGCGUCGCUUGACGCCCGCCAACAAUCAUCAAUGGCCGAAGAUUGCCAUAAUUUGCGAUGGCGUUAAUACCAUGAGGUCAGUUAGG------- .(((((((((.....((.(((.(((((.(((((....))))).))))).))).))...)))))))))....(((((........))))).((((((((....)))..))))).------- ( -41.80) >DroPer_CAF1 32137 113 - 1 ACCUGGCCAUCACACUGCUGGGCGGUGCUAGACGCCUGACGCCGGCCAACAAUCAUCAGUGGCCCAAGAUAGCCAUCAUUUGUGAUGGCGUCAAGAACAUGAGGCGAGCAGAG------- ..............((((((((((........)))))..(((((((((.(........))))))...(((.(((((((....))))))))))..........)))))))))..------- ( -43.90) >consensus AUCUGGCCAUCACACUGCUGGGCGGCGCUAGACGCUUGACGCCGGCCAACAAUCAUCAGUGGCCCAAGAUAGCCAUCAUUUGCGAUGGCGUCAAAAACAUGAGGUGAGUUAAC_______ ((((((((((..........(((((((.(((....))).)))).)))...........)))))).......((((((......))))))............))))............... (-25.20 = -25.45 + 0.25)

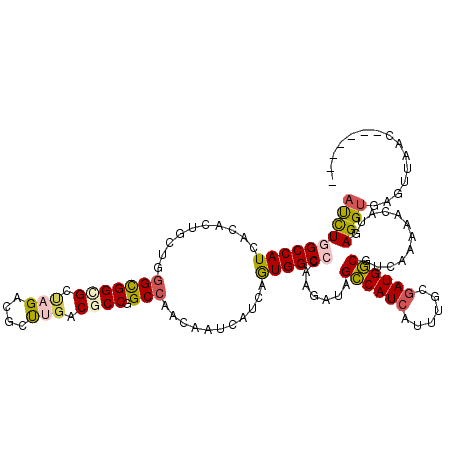
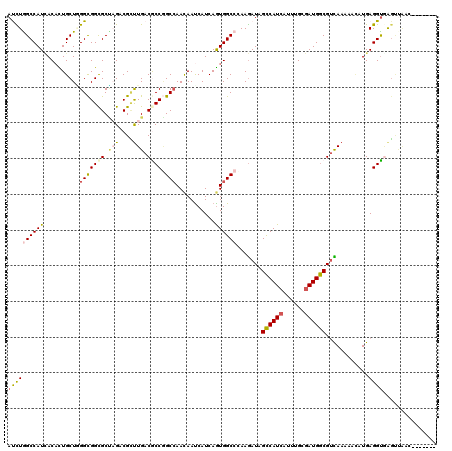
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:12 2006