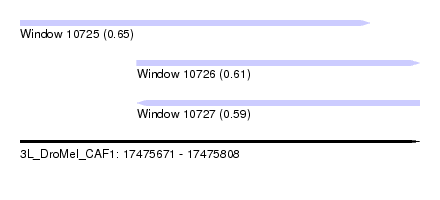
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,475,671 – 17,475,808 |
| Length | 137 |
| Max. P | 0.651671 |

| Location | 17,475,671 – 17,475,791 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17475671 120 + 23771897 CUGUGAUAAUAACUGCAAGAACUUCAUCUACUUUAUACUGAUCUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGCACACACCCCAA .((((.........(((((....(((............)))..))))).....((((((.........(((((.....)))))((((....)))).....))))))....))))...... ( -26.60) >DroVir_CAF1 40320 120 + 1 CUGUGAUAAUAACUGCAAGAACUUCAUCUACUUUAUACUGAUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUUAUGCGCUGCACACAUCCUAA .((((.........(((((((..(((............)))))))))).....((((((....((...(((((.....)))))((((....))))..)).))))))....))))...... ( -30.10) >DroGri_CAF1 39695 120 + 1 CUGUGAUAAUAACUGCAAGAACUUUAUCUACUUUAUAUUGAUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGCACACAUCCUAA .((((.........((((((((..(((.......)))..).))))))).....((((((.........(((((.....)))))((((....)))).....))))))....))))...... ( -29.30) >DroWil_CAF1 43935 120 + 1 CUGUGAUAAUAACUGCAAGAACUUCAUCUACUUUAUACUGAUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUUGUCAUGCGCUGUACACAUCCAAA .((((.........(((((((..(((............)))))))))).....((((((....(((..(((((.....)))))((((....)))).))).))))))....))))...... ( -29.10) >DroMoj_CAF1 39017 120 + 1 CUGUGAUAAUAACUGCAAGAACUUCAUCUACUUUAUAGCGAUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCUGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGUACACAUCCUAA .((((............(((......)))....((((((......((((...(((((....)))))...((((.....))))))))((((((.....).)))))))))))))))...... ( -31.50) >DroAna_CAF1 35271 120 + 1 CUGUGAUAAUAACUGCAAGAACUUCAUAUACUUUAUACUGAUCUUUGCCAUUUGCGUAUUCAUGCAUUCCACCUCCGAGGUGGGCAGUAUGCUUCUCGUCAUGCGGUGUACACAUCCCAA .((((.((...((((((.((.............(((((((............(((((....))))).((((((.....))))))))))))).......)).)))))).))))))...... ( -28.85) >consensus CUGUGAUAAUAACUGCAAGAACUUCAUCUACUUUAUACUGAUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGCACACAUCCUAA .((((.........((((((...(((............))).)))))).....((((((....((...(((((.....)))))((((....))))..)).))))))....))))...... (-25.10 = -25.80 + 0.70)

| Location | 17,475,711 – 17,475,808 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.42 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17475711 97 + 23771897 AUCUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGCACACACCCCAAAGGUAAUAA-------C-----CA-------GGAUU---- ....(((((...(((((....)))))..(((((.....)))))(((((..((..........)))))))............)))))...-------.-----..-------.....---- ( -25.60) >DroVir_CAF1 40360 91 + 1 AUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUUAUGCGCUGCACACAUCCUAAAGGUGAGU--------C-----AA---------------- ...((..((...(((((....)))))..(((((.....)))))(((((..((..........)))))))............))..)).--------.-----..---------------- ( -25.70) >DroGri_CAF1 39735 104 + 1 AUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGCACACAUCCUAAAGGUAACCAGACAUCCC-----CA-------UAUCU---- ....(((((...(((((....)))))..(((((.....)))))(((((..((..........)))))))............)))))...........-----..-------.....---- ( -25.50) >DroWil_CAF1 43975 101 + 1 AUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUUGUCAUGCGCUGUACACAUCCAAAGGGUAAUUA-------U-----UA-------UAUUCAUAC ....(((((...((.((((...(((((.(((((.....)))))((((....)))).....)))))..)))).)).......)))))...-------.-----..-------......... ( -26.30) >DroMoj_CAF1 39057 96 + 1 AUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCUGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGUACACAUCCUAAAGGUAAGU--------C-----CA-------AGCUC---- ...((((((...((.((((...(((((.(((((.....)))))((((....)))).....)))))..)))).)).......)))))).--------.-----..-------.....---- ( -25.80) >DroAna_CAF1 35311 109 + 1 AUCUUUGCCAUUUGCGUAUUCAUGCAUUCCACCUCCGAGGUGGGCAGUAUGCUUCUCGUCAUGCGGUGUACACAUCCCAAAGGUAGGAU-------ACUUUUCAGAGGAAGGGAAU---- (((((.(((....(((....(((((..((((((.....))))))..))))).....)))..((.((((....)))).))..))))))))-------.((((((....))))))...---- ( -29.50) >consensus AUUUUUGCCAUUUGCGUAUUUAUGCAUUCCACCUCCGAGGUGGGCAGCAUGCUGCUCGUCAUGCGCUGCACACAUCCUAAAGGUAAGUA_______C_____CA_______GGAUU____ ....(((((...(((((....)))))..(((((.....)))))(((((..((..........)))))))............))))).................................. (-23.92 = -23.42 + -0.50)
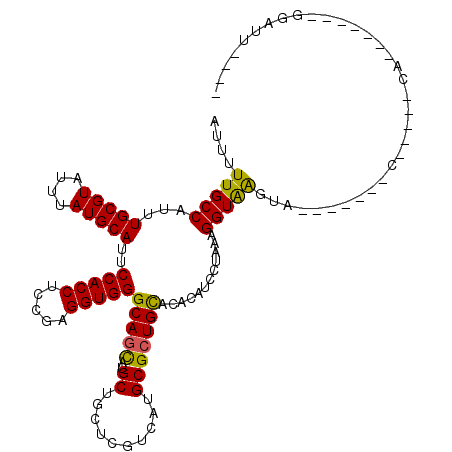
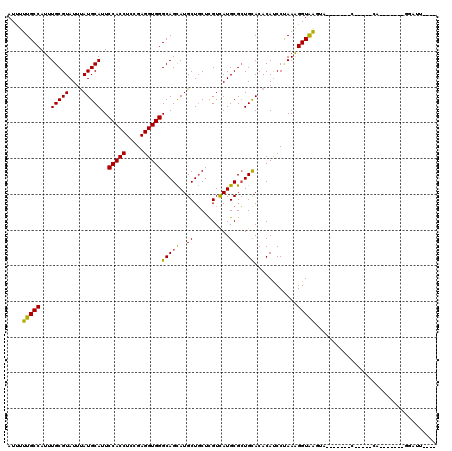
| Location | 17,475,711 – 17,475,808 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17475711 97 - 23771897 ----AAUCC-------UG-----G-------UUAUUACCUUUGGGGUGUGUGCAGCGCAUGACGAGCAGCAUGCUGCCCACCUCGGAGGUGGAAUGCAUAAAUACGCAAAUGGCAAAGAU ----.(((.-------..-----(-------(((((((((((((((((.(.(((((((.((.....))))..))))))))))))))))))....(((........)))))))))...))) ( -35.40) >DroVir_CAF1 40360 91 - 1 ----------------UU-----G--------ACUCACCUUUAGGAUGUGUGCAGCGCAUAACGAGCAGCAUGCUGCCCACCUCGGAGGUGGAAUGCAUAAAUACGCAAAUGGCAAAAAU ----------------..-----.--------..((((((((((((((((.....)))))...(.((((....)))).).))).))))))))..(((........)))............ ( -25.40) >DroGri_CAF1 39735 104 - 1 ----AGAUA-------UG-----GGGAUGUCUGGUUACCUUUAGGAUGUGUGCAGCGCAUGACGAGCAGCAUGCUGCCCACCUCGGAGGUGGAAUGCAUAAAUACGCAAAUGGCAAAAAU ----(((((-------(.-----...)))))).((((((....))...(((((((((((((.(.....)))))).))(((((.....)))))..))))))..........))))...... ( -28.50) >DroWil_CAF1 43975 101 - 1 GUAUGAAUA-------UA-----A-------UAAUUACCCUUUGGAUGUGUACAGCGCAUGACAAGCAGCAUGCUGCCCACCUCGGAGGUGGAAUGCAUAAAUACGCAAAUGGCAAAAAU ((((.....-------..-----.-------......((....)).((((((..(((((((........))))).))(((((.....)))))..)))))).))))((.....))...... ( -23.60) >DroMoj_CAF1 39057 96 - 1 ----GAGCU-------UG-----G--------ACUUACCUUUAGGAUGUGUACAGCGCAUGACGAGCAGCAUGCUGCCCACCUCAGAGGUGGAAUGCAUAAAUACGCAAAUGGCAAAAAU ----..(((-------((-----.--------.....((....))(((((.....)))))..)))))....((((..(((((.....)))))..(((........)))...))))..... ( -27.00) >DroAna_CAF1 35311 109 - 1 ----AUUCCCUUCCUCUGAAAAGU-------AUCCUACCUUUGGGAUGUGUACACCGCAUGACGAGAAGCAUACUGCCCACCUCGGAGGUGGAAUGCAUGAAUACGCAAAUGGCAAAGAU ----(((((...(((((((..(((-------((.((...((((..(((((.....)))))..)))).)).))))).......))))))).)))))..........((.....))...... ( -25.50) >consensus ____AAACA_______UG_____G_______UACUUACCUUUAGGAUGUGUACAGCGCAUGACGAGCAGCAUGCUGCCCACCUCGGAGGUGGAAUGCAUAAAUACGCAAAUGGCAAAAAU .....................................((....)).((((((....((((.....((((....))))(((((.....))))).)))).....))))))............ (-22.50 = -22.42 + -0.08)

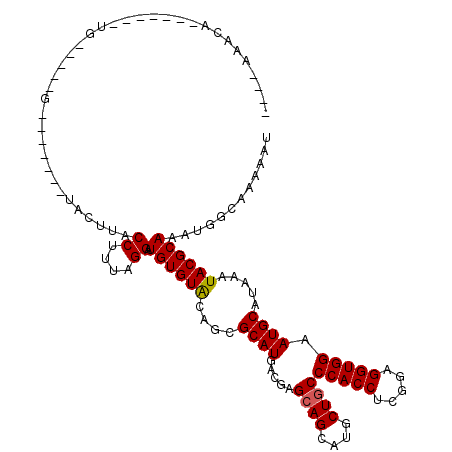
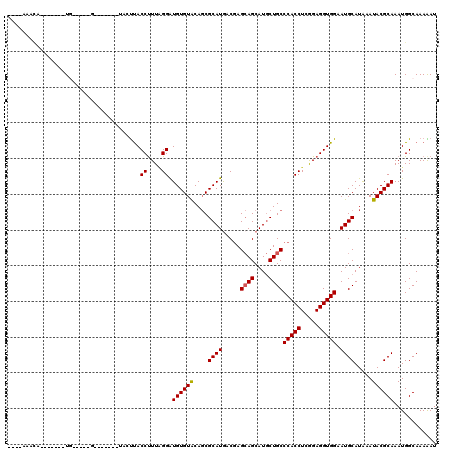
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:06 2006