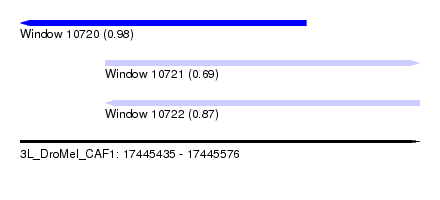
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,445,435 – 17,445,576 |
| Length | 141 |
| Max. P | 0.977157 |

| Location | 17,445,435 – 17,445,536 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -14.79 |
| Energy contribution | -14.71 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17445435 101 - 23771897 GAAAACACAAGUAGGUACAGGAAUAUGUUGUGAAUCGU--------AUUCUUGUGUGUACAUUGUGGUUUCACUGGGUUUUGUGGUCAGCUUAAAUUAGCAU--------AU--AUGUG ((((.((((((((..((((((((((((........)))--------)))))))))..))).))))).)))).................(((......)))..--------..--..... ( -28.40) >DroSec_CAF1 4686 110 - 1 GAAAACACAAGUAGGUACAGGAAUAGGUUGUGAAUCAU--------AUUCUUGUGUGUACAUUGUGGUUCCAUUGGGUUUUGUGGUCAGCUUAAAUUAGCACUAGUUGUUAUAA-AAUA .....((((((((..(((((((((((((.....))).)--------)))))))))..))).)))))..........(((((((((.(((((............)))))))))))-))). ( -28.50) >DroSim_CAF1 5165 102 - 1 GAAAACACAAGUAGGUACAGGAAUAGGUUGUGAAUCAU--------AUUCUUGUGUGUACAU-GUGGUUCCAUUGGGUUUUGUGGUCAGCUUAAAUUAGCAU--------AUACAUAUG .....((((.(((..(((((((((((((.....))).)--------)))))))))..))).)-)))...((((........))))...(((......)))..--------......... ( -25.00) >DroEre_CAF1 3776 100 - 1 AAAAACACAGGUAACUACAGGAAUAUGUUGUGAUUCGUACCCAUGUAUUCCUGUAUGUACAUUGUGUUUUCAAUGGGUUUUCUGGUCAGUUUAAAUUAUC------------------- .((((((((((((..(((((((((((((.(((.....)))..)))))))))))))..))).)))))))))..............................------------------- ( -25.50) >DroYak_CAF1 3768 92 - 1 GGAAACACAAGUAACUACAGGAAUAUGUUGUGAAUCGU--------AUUGCUGUGUGUACAUUGUGUUUUCAAUGGGUUUUCUGGUCAGUUUAAAUUAUC------------------- (((((((((((((..(((((.((((((........)))--------))).)))))..))).)))))))))).............................------------------- ( -25.80) >consensus GAAAACACAAGUAGGUACAGGAAUAUGUUGUGAAUCGU________AUUCUUGUGUGUACAUUGUGGUUUCAAUGGGUUUUGUGGUCAGCUUAAAUUAGCA_________AU_____U_ .....((((((((..(((((((((......................)))))))))..))).)))))...((((........)))).................................. (-14.79 = -14.71 + -0.08)


| Location | 17,445,465 – 17,445,576 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -17.37 |
| Energy contribution | -18.01 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17445465 111 + 23771897 AACCCAGUGAAACCACAAUGUACACACAAGAAU--------ACGAUUCACAACAUAUUCCUGUACCUACUUGUGUUUUCAUUGGACUUCUUGGUCGAAUGCGAAAUUUUCUUGGAGUGA ...(((((((((.(((((.(((...(((.((((--------(.(........).))))).)))...)))))))).)))))))))(((((..(((((....))).....))..))))).. ( -28.10) >DroSec_CAF1 4725 111 + 1 AACCCAAUGGAACCACAAUGUACACACAAGAAU--------AUGAUUCACAACCUAUUCCUGUACCUACUUGUGUUUUCAUUGGCGUUCUUGGUCGAACGCGAAAUUUUCUUGGACUGA ..((((((((((.(((((.(((...(((.((((--------(.(........).))))).)))...)))))))).))))))))((((((......))))))...........))..... ( -29.50) >DroSim_CAF1 5197 110 + 1 AACCCAAUGGAACCAC-AUGUACACACAAGAAU--------AUGAUUCACAACCUAUUCCUGUACCUACUUGUGUUUUCAUUGGCGUUCUUGGUCGAACGCGAAAUUUUCUUGGACUGA ..((((((((((.(((-(.(((...(((.((((--------(.(........).))))).)))...))).)))).))))))))((((((......))))))...........))..... ( -29.20) >DroEre_CAF1 3797 119 + 1 AACCCAUUGAAAACACAAUGUACAUACAGGAAUACAUGGGUACGAAUCACAACAUAUUCCUGUAGUUACCUGUGUUUUUAUUGGAGUUCUUGGUCGAAUGCGAAAUUGCCUUGGAGUGG ...(((.((((((((((..(((..((((((((((..(((((....))).))...))))))))))..))).)))))))))).)))(.(((..((.(((........)))))..))).).. ( -34.40) >DroYak_CAF1 3789 111 + 1 AACCCAUUGAAAACACAAUGUACACACAGCAAU--------ACGAUUCACAACAUAUUCCUGUAGUUACUUGUGUUUCCAUUAGAGUUCCUGGUCGAAUGCGACAUCGCCUUGGAUUGG .......((.((((((((.(((.((((((.(((--------(.(........).)))).)))).))))))))))))).)).........(..(((.((.(((....))).)).)))..) ( -25.70) >consensus AACCCAAUGAAACCACAAUGUACACACAAGAAU________ACGAUUCACAACAUAUUCCUGUACCUACUUGUGUUUUCAUUGGAGUUCUUGGUCGAAUGCGAAAUUUUCUUGGACUGA ...(((((((((.(((((.(((...(((.((((......................)))).)))...)))))))).))))))))).((((..(((((....))).....))..))))... (-17.37 = -18.01 + 0.64)


| Location | 17,445,465 – 17,445,576 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17445465 111 - 23771897 UCACUCCAAGAAAAUUUCGCAUUCGACCAAGAAGUCCAAUGAAAACACAAGUAGGUACAGGAAUAUGUUGUGAAUCGU--------AUUCUUGUGUGUACAUUGUGGUUUCACUGGGUU .....................(((......))).((((.(((((.((((((((..((((((((((((........)))--------)))))))))..))).))))).))))).)))).. ( -32.20) >DroSec_CAF1 4725 111 - 1 UCAGUCCAAGAAAAUUUCGCGUUCGACCAAGAACGCCAAUGAAAACACAAGUAGGUACAGGAAUAGGUUGUGAAUCAU--------AUUCUUGUGUGUACAUUGUGGUUCCAUUGGGUU .....((...........((((((......))))))(((((....((((((((..(((((((((((((.....))).)--------)))))))))..))).)))))....))))))).. ( -34.90) >DroSim_CAF1 5197 110 - 1 UCAGUCCAAGAAAAUUUCGCGUUCGACCAAGAACGCCAAUGAAAACACAAGUAGGUACAGGAAUAGGUUGUGAAUCAU--------AUUCUUGUGUGUACAU-GUGGUUCCAUUGGGUU .....((...........((((((......))))))(((((....((((.(((..(((((((((((((.....))).)--------)))))))))..))).)-)))....))))))).. ( -34.60) >DroEre_CAF1 3797 119 - 1 CCACUCCAAGGCAAUUUCGCAUUCGACCAAGAACUCCAAUAAAAACACAGGUAACUACAGGAAUAUGUUGUGAUUCGUACCCAUGUAUUCCUGUAUGUACAUUGUGUUUUCAAUGGGUU ..........((......))...........(((.(((.(.((((((((((((..(((((((((((((.(((.....)))..)))))))))))))..))).))))))))).).)))))) ( -31.30) >DroYak_CAF1 3789 111 - 1 CCAAUCCAAGGCGAUGUCGCAUUCGACCAGGAACUCUAAUGGAAACACAAGUAACUACAGGAAUAUGUUGUGAAUCGU--------AUUGCUGUGUGUACAUUGUGUUUUCAAUGGGUU ..((((((.(((((........))).))...........((((((((((((((..(((((.((((((........)))--------))).)))))..))).))))))))))).)))))) ( -36.90) >consensus UCACUCCAAGAAAAUUUCGCAUUCGACCAAGAACUCCAAUGAAAACACAAGUAGGUACAGGAAUAUGUUGUGAAUCGU________AUUCUUGUGUGUACAUUGUGGUUUCAAUGGGUU .....................(((......)))..(((.(((((.((((((((..(((((((((......................)))))))))..))).))))).))))).)))... (-19.95 = -20.15 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:01 2006