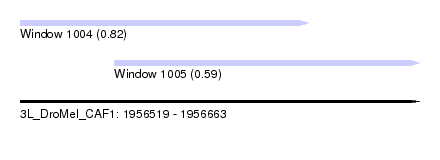
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,956,519 – 1,956,663 |
| Length | 144 |
| Max. P | 0.820023 |

| Location | 1,956,519 – 1,956,623 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 64.18 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.48 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
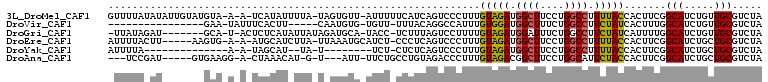
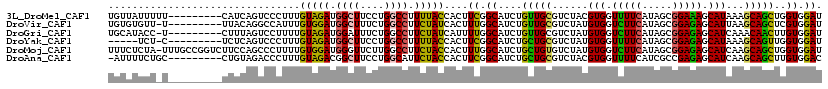
>3L_DroMel_CAF1 1956519 104 + 23771897 GUUUUAUAUAUUGUAUGUA-A-A-UCAUAUUUUA-UAGUGUU-AUUUUUCAUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUUUACCACUUCGGCAUCUGUUGCGUCUA ......((((..(((((..-.-.-.)))))..))-)).....-......((.(((.((....(((((.((((....)))).)))))......))...))).))...... ( -17.20) >DroVir_CAF1 7491 85 + 1 ----------------GAA-UAUUUCACUU-----CAAUGUG-UGUU-UUUACAGGCCAUUUGUGGAUGGCUUUCUGGCCUUCUAUCACUUUGGCAUCUGUUGCGUCUA ----------------...-.....(((..-----....)))-.((.-...((((((((..((((((.((((....)))).)))).))...))))..)))).))..... ( -20.80) >DroGri_CAF1 6353 97 + 1 -UUAUAGAU-------GCA-U-ACUCUCAUAUUAUAGAUGCA-UACC-UCUUUAGUCCUUUUGUAGAUGGAUUUCUGGCCUUCUAUCAUUUUGGCAUCUGUUGCGUCUA -...(((((-------(((-.-...........((((((((.-(((.-..............)))((((((..........))))))......)))))))))))))))) ( -21.86) >DroEre_CAF1 7681 99 + 1 AUUUUACUU-----AAGUG-A-A-AUGCAUCUUA-UUAAAUGCAUCU-CCCUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUUUACCACUUCGGCAUCUGCUGCGUCUA .........-----(((((-.-.-((((((....-....))))))..-..............(((((.((((....)))).))))))))))((((....))))...... ( -20.90) >DroYak_CAF1 7956 81 + 1 AUUUUA--------------A-A-UAGCAU--UA-U--------UCU-CUCUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUUUACCACUUCGGCAUCUGCUGCGUCUA ......--------------.-.-(((((.--..-.--------...-..............(((((.((((....)))).)))))..(....)....)))))...... ( -13.70) >DroAna_CAF1 6166 93 + 1 ---UCCGAU-----GUGAAGG-A-CUAAACAU-G-U---AUU-UUCUGCCUGUAGACCCUUUGUAGACGGCUUCCUGGCAUUCUACCACUUCGGCAUCUGCUGCGUCUA ---...(((-----((((((.-.-....(((.-(-(---(..-...))).))).........(((((..(((....)))..)))))..))))(((....)))))))).. ( -21.20) >consensus _UUUUA_AU_______GAA_A_A_UCACAU_UUA_U_AUGUG_UUCU_CCUUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUCUACCACUUCGGCAUCUGCUGCGUCUA ..............................................................(((((.((((....)))).))))).......(((.....)))..... (-10.76 = -10.48 + -0.28)
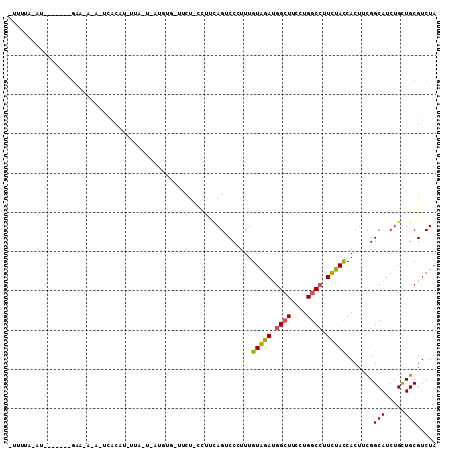
| Location | 1,956,553 – 1,956,663 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -23.39 |
| Energy contribution | -22.28 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
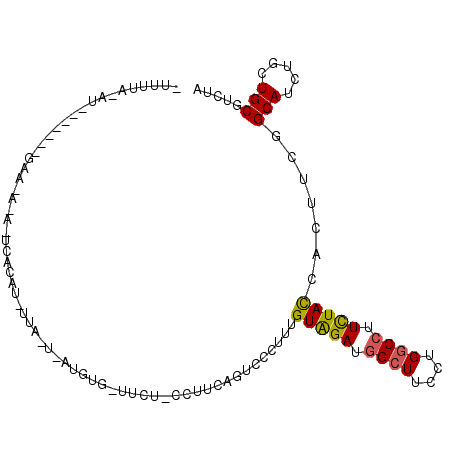
>3L_DroMel_CAF1 1956553 110 + 23771897 UGUUAUUUUU---------CAUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUUUACCACUUCGGCAUCUGUUGCGUCUACGUGGUUUUCAUAGCGGAAAGCAUAAAGCAGCUGGUGGAU ........((---------(((((((..((((((((((((((....)))).............(((.....)))))))))(((.(((((.....))))).)))))))..))))))))). ( -33.50) >DroVir_CAF1 7507 109 + 1 UGUGUGUU-U---------UUACAGGCCAUUUGUGGAUGGCUUUCUGGCCUUCUAUCACUUUGGCAUCUGUUGCGUCUAUGUGGUCUUCAUAGCGGAGAGCAUUAAGCAGCUCGUGGAU ........-(---------((((..((((..((((((.((((....)))).)))).))...))))....(((((......(((.(((((.....))))).)))...)))))..))))). ( -33.80) >DroGri_CAF1 6381 109 + 1 UGCAUACC-U---------CUUUAGUCCUUUUGUAGAUGGAUUUCUGGCCUUCUAUCAUUUUGGCAUCUGUUGCGUCUAUGUGGUCUUCAUAGCGGAGAGCAUCAAACAACUUGUGGAU ........-.---------.....((((....(((((((.(......(((............)))......).)))))))(((.(((((.....))))).)))............)))) ( -22.80) >DroYak_CAF1 7973 104 + 1 -----UCU-C---------UCUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUUUACCACUUCGGCAUCUGCUGCGUCUAUGUGGUUUUCAUAGCGGAGAGCAUAAAGCAGUUGGUGGAU -----...-.---------.............(((((.((((....)))).)))))..(....)((((.(((((.....((((.(((((.....))))).))))..))))).))))... ( -27.50) >DroMoj_CAF1 7318 118 + 1 UUUCUCUA-UUUGCCGGUCUUCCAGCCCUUUUGUGGAUGGGUUCUUGGCCUUCUACCACUUUGGCAUCUGCUGUGUCUAUGUGGUCUUCAUAGCGGAGAGCAUCAAGCAGCUGGUGGAU .......(-((..(((((...(((..((......)).)))((((((.((.....(((((...(((((.....)))))...))))).......)).))))))........)))))..))) ( -37.60) >DroAna_CAF1 6190 109 + 1 -AUUUUCUGC---------CUGUAGACCCUUUGUAGACGGCUUCCUGGCAUUCUACCACUUCGGCAUCUGCUGCGUCUACGUGGUUUUCAUCGCCGAGAGCAUCAAGCAGCUUGUGGAC -.......((---------((((((((.....(((((..(((....)))..))))).....((((....)))).))))))).)))........(((.((((........)))).))).. ( -30.40) >consensus UGUUUUCU_U_________CUUCAGUCCCUUUGUAGAUGGCUUCCUGGCCUUCUACCACUUCGGCAUCUGCUGCGUCUAUGUGGUCUUCAUAGCGGAGAGCAUCAAGCAGCUGGUGGAU ................................(((((.((((....)))).)))))....((.((....(((((......(((.(((((.....))))).)))...)))))..)).)). (-23.39 = -22.28 + -1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:49 2006