
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,438,733 – 17,438,870 |
| Length | 137 |
| Max. P | 0.999073 |

| Location | 17,438,733 – 17,438,844 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -20.15 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17438733 111 + 23771897 --AGCAACAUUUCGUAUUCGAAAUCCAUAAUUAAUUUGUUGACCUGCAACACAGUGCGUAUGUGUUAUAUUCAUUUAGAACAUUGCGCAUACGUCAUGUUGAGUCAUAG--AAUA --......((((((....)))))).........((((..((((...(((((....((((((((((.((.(((.....))).)).))))))))))..))))).))))..)--))). ( -27.80) >DroSec_CAF1 7636 111 + 1 AAAGCAACAUUUAGUAUUCGAAACACAUAAUUUAUUUGUUGACCUGUAACACAGUGCGUAUGUGUAAUAUUCUCUUAGAACAUUGCGCAUACGUCAUGUUCAG--ACAG--AAUA ....(((((..(((.................)))..)))))..((((...(((..(((((((((((((.(((.....))).)))))))))))))..)))....--))))--.... ( -29.53) >DroSim_CAF1 7356 111 + 1 --AGCAACAUUUCGUAUUCGAAAUCCAUAAUUAAUUUGUUGACCUGCAACACAUUGCGUAUGUGUAAUAUUCAUUUAGAACAUUGCGCAUACGUCAUGUUGAGUCACAG--AAUA --......((((((....))))))..........(((((.(((...(((((....(((((((((((((.(((.....))).)))))))))))))..))))).)))))))--)... ( -33.00) >DroEre_CAF1 7443 109 + 1 AAAACAUCA------GUUCAAAACCAAUAAUUUAUUUACUGACCAGCCGCACACUGCGUAUGUGUAAUAUUUAUAUACAACAUUGCCCAUACGCUAUGUUGAGUCACUGGUAAUA .......((------((....................))))(((((..(((((..(((((((.(((((.............))))).)))))))..)))...))..))))).... ( -22.77) >consensus __AGCAACAUUUCGUAUUCGAAACCCAUAAUUAAUUUGUUGACCUGCAACACAGUGCGUAUGUGUAAUAUUCAUUUAGAACAUUGCGCAUACGUCAUGUUGAGUCACAG__AAUA .......................................((((...(((((....(((((((((((((.(((.....))).)))))))))))))..))))).))))......... (-20.15 = -21.53 + 1.38)

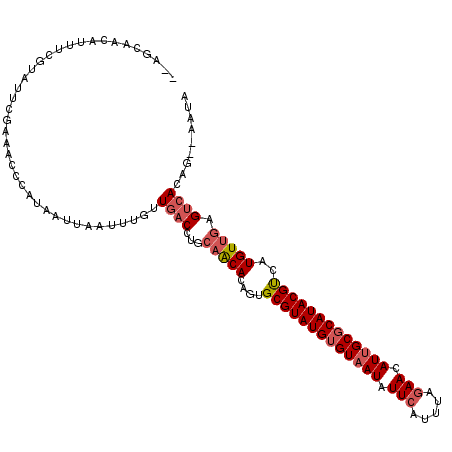
| Location | 17,438,733 – 17,438,844 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -17.06 |
| Energy contribution | -18.62 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17438733 111 - 23771897 UAUU--CUAUGACUCAACAUGACGUAUGCGCAAUGUUCUAAAUGAAUAUAACACAUACGCACUGUGUUGCAGGUCAACAAAUUAAUUAUGGAUUUCGAAUACGAAAUGUUGCU-- ....--...((((((((((((.((((((.(..((((((.....))))))..).))))))...)))))))..)))))...............((((((....))))))......-- ( -25.70) >DroSec_CAF1 7636 111 - 1 UAUU--CUGU--CUGAACAUGACGUAUGCGCAAUGUUCUAAGAGAAUAUUACACAUACGCACUGUGUUACAGGUCAACAAAUAAAUUAUGUGUUUCGAAUACUAAAUGUUGCUUU ...(--((((--....(((((.((((((.(.(((((((.....))))))).).)))))))).)))...))))).(((((......(((.((((.....))))))).))))).... ( -22.20) >DroSim_CAF1 7356 111 - 1 UAUU--CUGUGACUCAACAUGACGUAUGCGCAAUGUUCUAAAUGAAUAUUACACAUACGCAAUGUGUUGCAGGUCAACAAAUUAAUUAUGGAUUUCGAAUACGAAAUGUUGCU-- ...(--(((..((...((((..((((((.(.(((((((.....))))))).).))))))..))))))..))))..................((((((....))))))......-- ( -29.30) >DroEre_CAF1 7443 109 - 1 UAUUACCAGUGACUCAACAUAGCGUAUGGGCAAUGUUGUAUAUAAAUAUUACACAUACGCAGUGUGCGGCUGGUCAGUAAAUAAAUUAUUGGUUUUGAAC------UGAUGUUUU ((((((((((......((((.(((((((.(.((((((.......)))))).).))))))).))))...)))))).)))).......(((..((.....))------..))).... ( -28.30) >consensus UAUU__CUGUGACUCAACAUGACGUAUGCGCAAUGUUCUAAAUGAAUAUUACACAUACGCACUGUGUUGCAGGUCAACAAAUAAAUUAUGGAUUUCGAAUACGAAAUGUUGCU__ .........((((((((((((.((((((.(.(((((((.....))))))).).))))))...)))))))..)))))....................................... (-17.06 = -18.62 + 1.56)

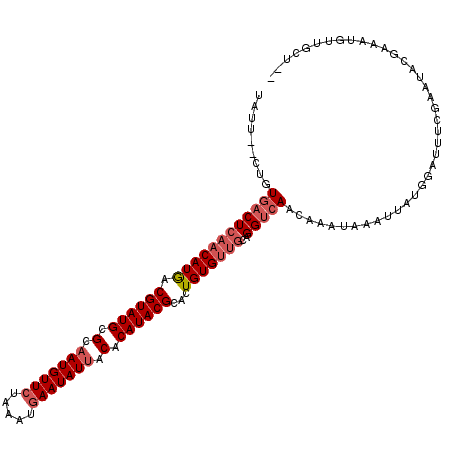

| Location | 17,438,766 – 17,438,870 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17438766 104 + 23771897 UUGUUGACCUGCAACACAGUGCGUAUGUGUUAUAUUCAUUUAGAACAUUGCGCAUACGUCAUGUUGAGUCAUAG--AAUAAAUGAUUGCUUAAAUUAAAGGAAUAA-------------- ....((((...(((((....((((((((((.((.(((.....))).)).))))))))))..))))).))))...--........(((.(((......))).)))..-------------- ( -24.90) >DroSec_CAF1 7671 102 + 1 UUGUUGACCUGUAACACAGUGCGUAUGUGUAAUAUUCUCUUAGAACAUUGCGCAUACGUCAUGUUCAG--ACAG--AAUAAUUGAUUGCUUAAAUUAAAGGAAUAA-------------- ........((((...(((..(((((((((((((.(((.....))).)))))))))))))..)))....--))))--........(((.(((......))).)))..-------------- ( -27.30) >DroSim_CAF1 7389 104 + 1 UUGUUGACCUGCAACACAUUGCGUAUGUGUAAUAUUCAUUUAGAACAUUGCGCAUACGUCAUGUUGAGUCACAG--AAUAAUUGAUUGCUUAAAUUAAAGGAAUAA-------------- ((((.(((...(((((....(((((((((((((.(((.....))).)))))))))))))..))))).)))))))--........(((.(((......))).)))..-------------- ( -29.60) >DroEre_CAF1 7472 120 + 1 UUACUGACCAGCCGCACACUGCGUAUGUGUAAUAUUUAUAUACAACAUUGCCCAUACGCUAUGUUGAGUCACUGGUAAUAAAAUAUUGCCUUAAUUAAGGGAUUGUUCCACUUUUAUAUA ....((((((((........(((((((.(((((.............))))).)))))))...)))).))))..(((((((...)))))))......((((((....))).)))....... ( -26.72) >DroYak_CAF1 7599 116 + 1 ---GUGUCCAGGGGCACACUGCGUAUGUGUAAUAUUUAUAGUGAACAUUGCUCAUACGUCGUGUUGAGUCACGGAUAAUAAAAGAUGGCUCUAGUUAAAAGAUUAUUCUUCUUUUGAAU- ---(((((....)))))((.(((((((.(((((.(((.....))).))))).))))))).))...(((((((...........).))))))...((((((((.......))))))))..- ( -32.70) >consensus UUGUUGACCUGCAACACACUGCGUAUGUGUAAUAUUCAUUUAGAACAUUGCGCAUACGUCAUGUUGAGUCACAG__AAUAAAUGAUUGCUUAAAUUAAAGGAAUAA______________ ....((((...(((((....(((((((((((((.(((.....))).)))))))))))))..))))).))))................................................. (-18.24 = -19.60 + 1.36)

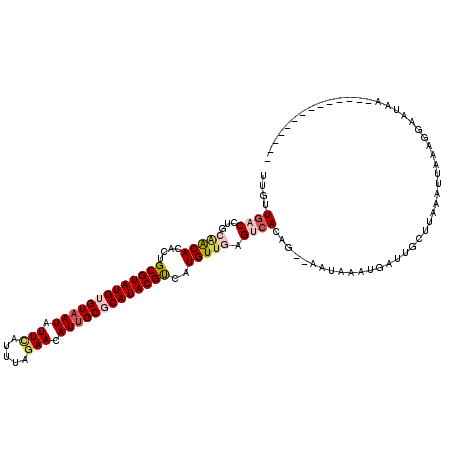

| Location | 17,438,766 – 17,438,870 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -15.08 |
| Energy contribution | -17.16 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17438766 104 - 23771897 --------------UUAUUCCUUUAAUUUAAGCAAUCAUUUAUU--CUAUGACUCAACAUGACGUAUGCGCAAUGUUCUAAAUGAAUAUAACACAUACGCACUGUGUUGCAGGUCAACAA --------------..............................--...((((((((((((.((((((.(..((((((.....))))))..).))))))...)))))))..))))).... ( -21.90) >DroSec_CAF1 7671 102 - 1 --------------UUAUUCCUUUAAUUUAAGCAAUCAAUUAUU--CUGU--CUGAACAUGACGUAUGCGCAAUGUUCUAAGAGAAUAUUACACAUACGCACUGUGUUACAGGUCAACAA --------------.............................(--((((--....(((((.((((((.(.(((((((.....))))))).).)))))))).)))...)))))....... ( -20.70) >DroSim_CAF1 7389 104 - 1 --------------UUAUUCCUUUAAUUUAAGCAAUCAAUUAUU--CUGUGACUCAACAUGACGUAUGCGCAAUGUUCUAAAUGAAUAUUACACAUACGCAAUGUGUUGCAGGUCAACAA --------------.............................(--(((..((...((((..((((((.(.(((((((.....))))))).).))))))..))))))..))))....... ( -25.50) >DroEre_CAF1 7472 120 - 1 UAUAUAAAAGUGGAACAAUCCCUUAAUUAAGGCAAUAUUUUAUUACCAGUGACUCAACAUAGCGUAUGGGCAAUGUUGUAUAUAAAUAUUACACAUACGCAGUGUGCGGCUGGUCAGUAA .......(((.(((....))))))...............(((((((((((......((((.(((((((.(.((((((.......)))))).).))))))).))))...)))))).))))) ( -29.20) >DroYak_CAF1 7599 116 - 1 -AUUCAAAAGAAGAAUAAUCUUUUAACUAGAGCCAUCUUUUAUUAUCCGUGACUCAACACGACGUAUGAGCAAUGUUCACUAUAAAUAUUACACAUACGCAGUGUGCCCCUGGACAC--- -.......((((((....(((.......)))....))))))....((((.(.....((((..((((((.(.((((((.......)))))).).))))))..))))...).))))...--- ( -22.10) >consensus ______________UUAUUCCUUUAAUUUAAGCAAUCAUUUAUU__CUGUGACUCAACAUGACGUAUGCGCAAUGUUCUAAAUGAAUAUUACACAUACGCACUGUGUUGCAGGUCAACAA .................................................((((((((((((.((((((.(.(((((((.....))))))).).))))))...)))))))..))))).... (-15.08 = -17.16 + 2.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:54 2006