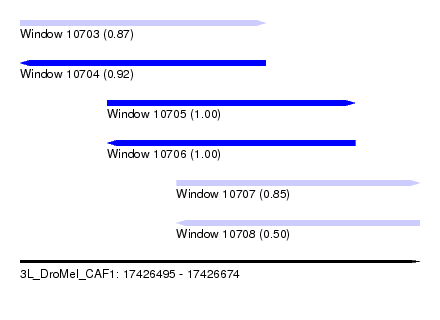
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,426,495 – 17,426,674 |
| Length | 179 |
| Max. P | 0.999510 |

| Location | 17,426,495 – 17,426,605 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.37 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -8.23 |
| Energy contribution | -7.79 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17426495 110 + 23771897 ACUAACAAGCUGAUAAUAAAUUGAAAAAUCUAUCAACGGUAGCAGCC-CCGCUAUUCCGUGGGGGGGGGGGGGGGUUGCUGAUGUUCUGAUGGGUAACUAUCAGCUUGCUG .....((((((((((......(....)(((((((((((.((((((((-((.((.((((.....)))).)).)))))))))).)))..))))))))...))))))))))... ( -47.20) >DroVir_CAF1 38368 92 + 1 CUACACAGACAUAGUAUACAGCAUAUCUACAGAGAAAUAGUCAUGUG-AAAAUCAA----AUGCUC--A-A-GAAGC---------UGCCACAAAUACGCUCUGGAGUCG- ...(((((((...((.....))...(((....)))....))).))))-........----..((((--.-(-((.((---------............))))).))))..- ( -14.20) >DroSec_CAF1 33880 101 + 1 ACUGACAAGCUGAUAAUAAAUUGAAAAAUCUAUCAACGGAAGCAGCC-CCACUUUUC-GCGGGGGGGAGGG-GGGUU-------UUCUGAUGGGUAACUAUCAGCUAGCUG .......((((((((......(....)((((((((..((((((..((-((.(((..(-.....)..)))))-)))))-------)))))))))))...))))))))..... ( -33.70) >DroSim_CAF1 33494 99 + 1 ACUGACAAGCUGAUAAUAAAUUGAAAAAUCUAUCAACGGUAGCAGCC-CCACUUUUC-GCGGGGGG--AGG-GGGUU-------UUCUGAUGGGUAACUAUCAGCUUGCUG .....((((((((((......(....)((((((((..((....((((-((.(((..(-....)..)--)))-)))))-------.))))))))))...))))))))))... ( -33.70) >DroEre_CAF1 34726 85 + 1 ACUGACAAGCUGAUAAUAAAUUGAAAAAUCUAUCAACGUUAGCAGCCCCCACUUUU--------------G-GGG-----------UUGAUAGGGAGCUAUCAGUUUGCUG .....((((((((((.....((((........))))(.(((.(((((((.......--------------)-)))-----------))).))).)...))))))))))... ( -25.60) >DroYak_CAF1 33827 85 + 1 ACUGACAAGCUGAUAAUAAAUUGAAAAAUCUAUCAACGGUAGCAGCCGCCACUUUG--------------G-GGG-----------CUGAUGGGGAAAUAUCCGCUGGCUG .......((((((((....(((....))).))))).((((..(((((.((.....)--------------)-.))-----------)))....(((....)))))))))). ( -23.70) >consensus ACUGACAAGCUGAUAAUAAAUUGAAAAAUCUAUCAACGGUAGCAGCC_CCACUUUU____GGGGGG____G_GGGUU_________CUGAUGGGUAACUAUCAGCUUGCUG ........((((........((((........))))......))))........................................(((((((....)))))))....... ( -8.23 = -7.79 + -0.44)


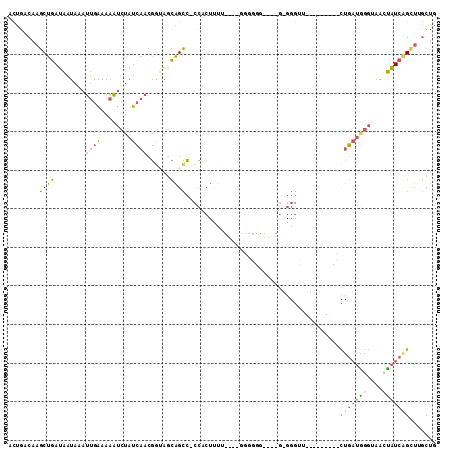
| Location | 17,426,495 – 17,426,605 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 61.37 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -8.73 |
| Energy contribution | -9.65 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17426495 110 - 23771897 CAGCAAGCUGAUAGUUACCCAUCAGAACAUCAGCAACCCCCCCCCCCCCCCACGGAAUAGCGG-GGCUGCUACCGUUGAUAGAUUUUUCAAUUUAUUAUCAGCUUGUUAGU .((((((((((((((................((((.(((((....((......))....).))-)).))))...(((((........)))))..))))))))))))))... ( -33.40) >DroVir_CAF1 38368 92 - 1 -CGACUCCAGAGCGUAUUUGUGGCA---------GCUUC-U-U--GAGCAU----UUGAUUUU-CACAUGACUAUUUCUCUGUAGAUAUGCUGUAUACUAUGUCUGUGUAG -.........((((((((((..(.(---------(....-.-.--.(((((----.((.....-)).))).))....)))..)))))))))).(((((.......))))). ( -18.60) >DroSec_CAF1 33880 101 - 1 CAGCUAGCUGAUAGUUACCCAUCAGAA-------AACCC-CCCUCCCCCCCGC-GAAAAGUGG-GGCUGCUUCCGUUGAUAGAUUUUUCAAUUUAUUAUCAGCUUGUCAGU ..((.((((((((((............-------.....-.......((((((-.....))))-))........(((((........)))))..)))))))))).)).... ( -26.30) >DroSim_CAF1 33494 99 - 1 CAGCAAGCUGAUAGUUACCCAUCAGAA-------AACCC-CCU--CCCCCCGC-GAAAAGUGG-GGCUGCUACCGUUGAUAGAUUUUUCAAUUUAUUAUCAGCUUGUCAGU ..(((((((((((((............-------.....-...--..((((((-.....))))-))........(((((........)))))..))))))))))))).... ( -30.40) >DroEre_CAF1 34726 85 - 1 CAGCAAACUGAUAGCUCCCUAUCAA-----------CCC-C--------------AAAAGUGGGGGCUGCUAACGUUGAUAGAUUUUUCAAUUUAUUAUCAGCUUGUCAGU ......((((((((...........-----------(((-(--------------(....)))))((((.(((.(((((........)))))...))).)))))))))))) ( -23.70) >DroYak_CAF1 33827 85 - 1 CAGCCAGCGGAUAUUUCCCCAUCAG-----------CCC-C--------------CAAAGUGGCGGCUGCUACCGUUGAUAGAUUUUUCAAUUUAUUAUCAGCUUGUCAGU ..((.((((((....)))....(((-----------((.-(--------------(.....)).))))).....(.(((((((((....))))))))).).))).)).... ( -20.50) >consensus CAGCAAGCUGAUAGUUACCCAUCAG_________AACCC_C____CCCCCC____AAAAGUGG_GGCUGCUACCGUUGAUAGAUUUUUCAAUUUAUUAUCAGCUUGUCAGU ..((.((((((((((...........................................(((((...)))))...(((((........)))))..)))))))))).)).... ( -8.73 = -9.65 + 0.92)
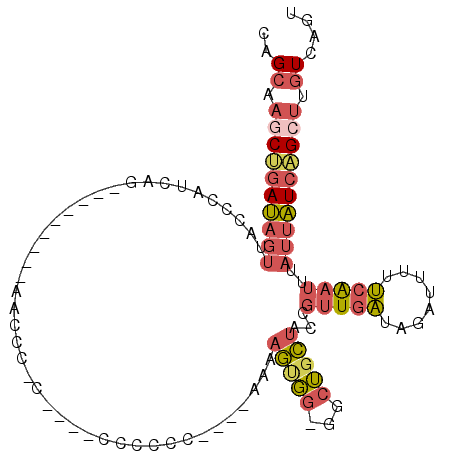

| Location | 17,426,534 – 17,426,645 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -22.71 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17426534 111 + 23771897 UAGCAGCC-CCGCUAUUCCGUGGGGGGGGGGGGGGGUUGCUGAUGUUCUGAUGGGUAACUAUCAGCUUGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACA ((((((((-((.((.((((.....)))).)).))))))))))..(((.((((......((((((((.....)))))))).......)))).)))((((((.....)))))). ( -48.92) >DroSec_CAF1 33919 102 + 1 AAGCAGCC-CCACUUUUC-GCGGGGGGGAGGG-GGGUU-------UUCUGAUGGGUAACUAUCAGCUAGCUGUUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACA .(((((((-((.(((..(-.....)..))).)-)))))-------..(((((((....)))))))...)))..((((.........))))...(((((((.....))))))) ( -37.20) >DroSim_CAF1 33533 100 + 1 UAGCAGCC-CCACUUUUC-GCGGGGGG--AGG-GGGUU-------UUCUGAUGGGUAACUAUCAGCUUGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACA ((((((((-((.(((..(-....)..)--)))-))...-------..(((((((....)))))))...)))))))..................(((((((.....))))))) ( -38.80) >DroEre_CAF1 34765 86 + 1 UAGCAGCCCCCACUUUU--------------G-GGG-----------UUGAUAGGGAGCUAUCAGUUUGCUGCUGAUGGAGUAGAAAUCAUAAUAUCAGGAUUGACCUGACA ((((((((((((....)--------------)-)))-----------(((((((....)))))))...)))))))....................(((((.....))))).. ( -31.00) >DroYak_CAF1 33866 86 + 1 UAGCAGCCGCCACUUUG--------------G-GGG-----------CUGAUGGGGAAAUAUCCGCUGGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACA (((((((((((......--------------.-.))-----------)....(.(((....))).).))))))))..................(((((((.....))))))) ( -31.60) >consensus UAGCAGCC_CCACUUUUC_G_GGGGGG___GG_GGGUU_______UUCUGAUGGGUAACUAUCAGCUUGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACA (((((((..((.(..................).))............(((((((....)))))))...)))))))..................(((((((.....))))))) (-22.71 = -23.03 + 0.32)

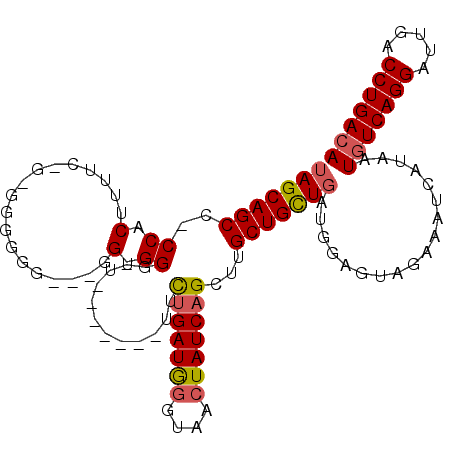

| Location | 17,426,534 – 17,426,645 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17426534 111 - 23771897 UGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCAAGCUGAUAGUUACCCAUCAGAACAUCAGCAACCCCCCCCCCCCCCCACGGAAUAGCGG-GGCUGCUA (((((((.....)))))))...................((((((..((((((.(((.........)))))))))..............(((.(......).))-))))))). ( -34.90) >DroSec_CAF1 33919 102 - 1 UGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAACAGCUAGCUGAUAGUUACCCAUCAGAA-------AACCC-CCCUCCCCCCCGC-GAAAAGUGG-GGCUGCUU (((((((.....))))))).....................((((...(((((........)))))..-------.....-........(((((-.....))))-)))))... ( -27.70) >DroSim_CAF1 33533 100 - 1 UGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCAAGCUGAUAGUUACCCAUCAGAA-------AACCC-CCU--CCCCCCGC-GAAAAGUGG-GGCUGCUA (((((((.....)))))))...................((((((...(((((........)))))..-------.....-...--...(((((-.....))))-))))))). ( -33.30) >DroEre_CAF1 34765 86 - 1 UGUCAGGUCAAUCCUGAUAUUAUGAUUUCUACUCCAUCAGCAGCAAACUGAUAGCUCCCUAUCAA-----------CCC-C--------------AAAAGUGGGGGCUGCUA (((((((.....)))))))...................((((((....((((((....)))))).-----------(((-(--------------(....))))))))))). ( -32.00) >DroYak_CAF1 33866 86 - 1 UGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCCAGCGGAUAUUUCCCCAUCAG-----------CCC-C--------------CAAAGUGGCGGCUGCUA (((((((.....)))))))...................(((((((.(.(((....))).)....(-----------((.-.--------------......)))))))))). ( -30.90) >consensus UGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCAAGCUGAUAGUUACCCAUCAGAA_______AACCC_CC___CCCCCC_C_GAAAAGUGG_GGCUGCUA (((((((.....)))))))...................((((((...(((((........)))))...........((.......................))..)))))). (-17.84 = -18.28 + 0.44)

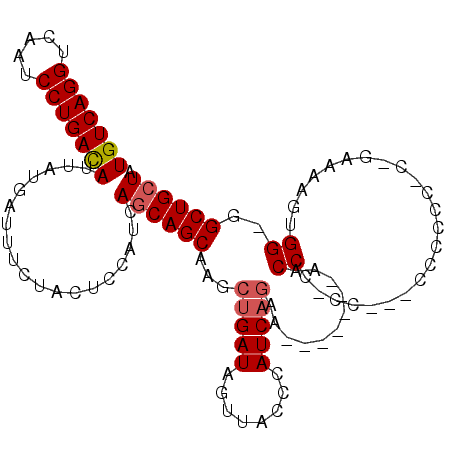
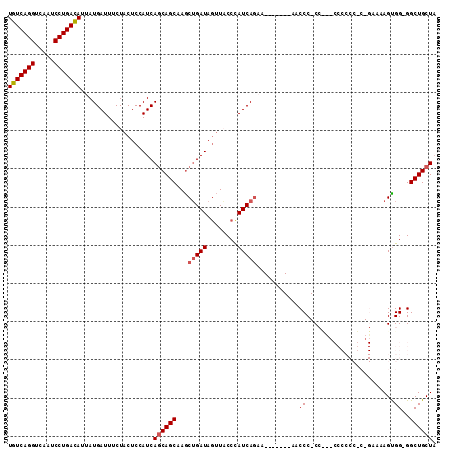
| Location | 17,426,565 – 17,426,674 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17426565 109 + 23771897 GGGGUUGCUGAUGUUCUGAUGGGUAACUAUCAGCUUGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACAGCUA----------CAAAAACAAUACGAACAUCCAGCUU ......((((((((((((.((.....((((((((.....)))))))).((((.........(((((((.....))))))).)))----------).....)).)).)))))).)))).. ( -38.40) >DroPse_CAF1 35904 100 + 1 -AGG-----------CGGAGAGGAAA--A-----UCCAUGCUGAUGAAGUGAAAAUCAUAAUGUCAGGAUGGACUUGACAGCUAAUGGCGGCAGCAAAAACAAUAUCCACAUCCUGCCG -.((-----------((((..(((..--.-----(((((.((((((..(((.....)))..)))))).))))).(((...(((......)))..)))........)))...))).))). ( -30.40) >DroSec_CAF1 33949 101 + 1 -GGGUU-------UUCUGAUGGGUAACUAUCAGCUAGCUGUUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACAGCUA----------CAAAAACAAUACGAACAUCCUGCGG -.((((-------..(((((((....)))))))..))))((.((((..(((..........(((((((.....)))))))....----------.........)))...))))..)).. ( -29.00) >DroSim_CAF1 33561 101 + 1 -GGGUU-------UUCUGAUGGGUAACUAUCAGCUUGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACAGCUA----------CAAAAACAAUACGAACAUCCUGCGG -((((.-------(((((.((.....((((((((.....)))))))).((((.........(((((((.....))))))).)))----------).....)).)).))).))))..... ( -31.30) >DroEre_CAF1 34783 97 + 1 -GGG-----------UUGAUAGGGAGCUAUCAGUUUGCUGCUGAUGGAGUAGAAAUCAUAAUAUCAGGAUUGACCUGACAGCUA----------CAAAAACAAUGCCAACAUCCUGCGG -.((-----------(((((((....)))))).((((..((((((((........))))....(((((.....)))))))))..----------))))......)))............ ( -23.40) >DroYak_CAF1 33884 97 + 1 -GGG-----------CUGAUGGGGAAAUAUCCGCUGGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACAGCUG----------CAAAAACAAUACCAACAUCCUGCGA -(((-----------.((.((((((....)))((.(((((((.....))))..........(((((((.....)))))))))))----------)..........))).)).))).... ( -26.90) >consensus _GGG___________CUGAUGGGUAACUAUCAGCUUGCUGCUGAUGGAGUAGAAAUCAUAAUGUCAGGAUUGACCUGACAGCUA__________CAAAAACAAUACCAACAUCCUGCGG ...............(((((((....)))))))......((.((((..((((....)....(((((((.....))))))).......................)))...))))..)).. (-16.75 = -17.67 + 0.92)

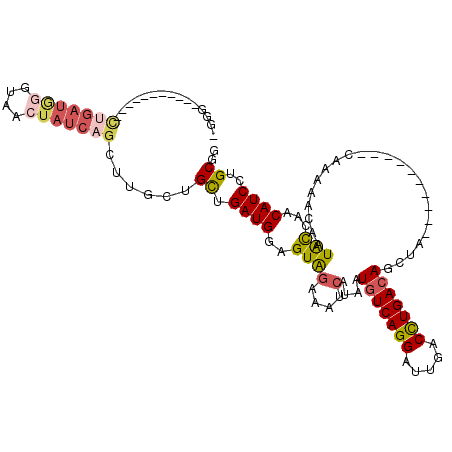

| Location | 17,426,565 – 17,426,674 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -13.13 |
| Energy contribution | -14.43 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17426565 109 - 23771897 AAGCUGGAUGUUCGUAUUGUUUUUG----------UAGCUGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCAAGCUGAUAGUUACCCAUCAGAACAUCAGCAACCCC ..((((.((((((....((.....(----------(((((((((((.....)))))).................((((((.....)))))))))))).))...))))))))))...... ( -35.00) >DroPse_CAF1 35904 100 - 1 CGGCAGGAUGUGGAUAUUGUUUUUGCUGCCGCCAUUAGCUGUCAAGUCCAUCCUGACAUUAUGAUUUUCACUUCAUCAGCAUGGA-----U--UUUCCUCUCCG-----------CCU- .(((.(((.(.(((..........((((.......))))....((((((((.((((.....(((........))))))).)))))-----)--))))).)))))-----------)).- ( -26.70) >DroSec_CAF1 33949 101 - 1 CCGCAGGAUGUUCGUAUUGUUUUUG----------UAGCUGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAACAGCUAGCUGAUAGUUACCCAUCAGAA-------AACCC- ..((..((((...(((......(..----------(((.(((((((.....))))))))))..)....)))..))))....))...(((((........)))))..-------.....- ( -21.40) >DroSim_CAF1 33561 101 - 1 CCGCAGGAUGUUCGUAUUGUUUUUG----------UAGCUGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCAAGCUGAUAGUUACCCAUCAGAA-------AACCC- ......((((...(((......(..----------(((.(((((((.....))))))))))..)....)))...((((((.....)))))).......))))....-------.....- ( -24.00) >DroEre_CAF1 34783 97 - 1 CCGCAGGAUGUUGGCAUUGUUUUUG----------UAGCUGUCAGGUCAAUCCUGAUAUUAUGAUUUCUACUCCAUCAGCAGCAAACUGAUAGCUCCCUAUCAA-----------CCC- .....((.((((((....((..(..----------(((.(((((((.....))))))))))..).....))....))))))......((((((....)))))).-----------)).- ( -21.60) >DroYak_CAF1 33884 97 - 1 UCGCAGGAUGUUGGUAUUGUUUUUG----------CAGCUGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCCAGCGGAUAUUUCCCCAUCAG-----------CCC- ..((.((.(((((((...((.....----------((..(((((((.....)))))))...))......))...))))))).)).))(((....))).......-----------...- ( -26.80) >consensus CCGCAGGAUGUUCGUAUUGUUUUUG__________UAGCUGUCAGGUCAAUCCUGACAUUAUGAUUUCUACUCCAUCAGCAGCAAGCUGAUAGUUACCCAUCAG___________CCC_ ......((((.............................(((((((.....)))))))................((((((.....)))))).......))))................. (-13.13 = -14.43 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:48 2006