
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,414,611 – 17,414,725 |
| Length | 114 |
| Max. P | 0.721565 |

| Location | 17,414,611 – 17,414,725 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17414611 114 - 23771897 GAGAGCAGUGUCCUCGCUGCUCAGAUCUGGCGGUGGGGUGGCAUAUAUGGAUGCAGCCAUUUUGGAUGAUAUUACUGCGG--GG--CG-GGGGAAGAAAGACGGAAAG-GGAGAAACAUU ..((((((((....)))))))).(.(((.((((((((((.((((......)))).)))(((......))).))))))).)--))--).-...................-........... ( -32.10) >DroVir_CAF1 22442 117 - 1 GAGCGCUGUGUCCUCGCUGCUCAGAUCCGGCGGUGGGGUGGCAUAAAUGGAUGCGGCCAUUUUGGAUGAUAUUACUGUGAGAAUGACA-GCGACAACAAAAAGCAGAA-AAGAAAGCAU- ....(((.(((..((((((.(((..((..((((((((((.((((......)))).)))(((......))).)))))))..)).)))))-))))..)))...)))....-..........- ( -36.10) >DroPse_CAF1 22260 116 - 1 GAGAGCAGUGUCCUCGCUGCUCAGAUCUGGCGGUGGGGUGGCAUAUAUGGAUGCAGCCAUUUUGGAUGAUAUUACUGCAAAAGA--GA-GAGGGAGAGAGAGGAGAAA-GGGGAGAAAUG ..((((((((....))))))))...(((.((((((((((.((((......)))).)))(((......))).)))))))...)))--..-...................-........... ( -30.80) >DroGri_CAF1 21023 114 - 1 GAGCGCAGUGUCUUCGCUGCUCAGAUCUGGCGGUGGGGUGGCAUAUAUGGAUGCGGCCAUUUUGGAUGAUAUUACUGUGUACAAAAUAUACGAAAACAACAUGCAGAG-AAAA----AU- (.((((((((...(((((((.((....)))))))))(((.((((......)))).)))..............)))))))).)..........................-....----..- ( -30.40) >DroMoj_CAF1 23045 114 - 1 GAGCGCUGUAUCCUCGCUGCUCAGAUCCGGCGGUGGGGUGGCAUAUAUGGAUGCGGCCAUUUUGGAUGAUAUUACUGUGAAUGGAACG-G----AACAAGAGGAAGAACGACAAAGCAU- ..((..(((.(((((..((.((.(.((((((((((((((.((((......)))).)))(((......))).)))))))...)))).).-)----).)).)))))......)))..))..- ( -31.60) >DroAna_CAF1 22354 102 - 1 GAGAGCAGUGUCCUCGCUGCUCAGAUCUGGCGGUGGGGUGGCAUAUAUGGAUGCAGCCAUUUUGGAUGAUAUUACUAGAA--GG-----GGCGA----------CAAA-AGAGACUCAUU (((.((.....(((((((((.((....)))))))))))..)).........((..(((.((((((.........))))))--..-----)))..----------))..-.....)))... ( -30.70) >consensus GAGAGCAGUGUCCUCGCUGCUCAGAUCUGGCGGUGGGGUGGCAUAUAUGGAUGCAGCCAUUUUGGAUGAUAUUACUGUGAA_GG__CA_GCGGAAACAAGAGGAAAAA_AAAAAAACAU_ ....(((((((((((((((((.......))))))))(((.((((......)))).))).....))))......))))).......................................... (-23.63 = -23.85 + 0.22)

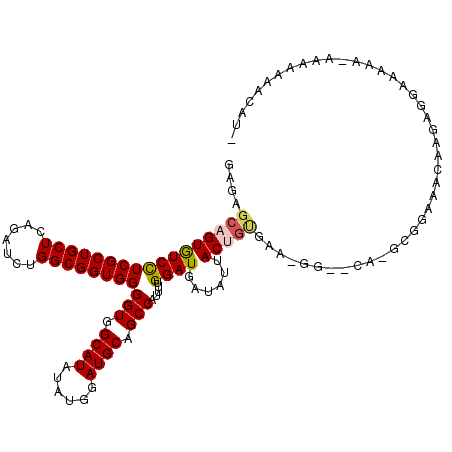

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:42 2006