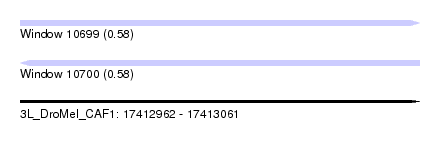
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,412,962 – 17,413,061 |
| Length | 99 |
| Max. P | 0.578774 |

| Location | 17,412,962 – 17,413,061 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17412962 99 + 23771897 UGUAGCUGGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUU---C---------UUUUU-CCUGCUCUCUGCCCGCCAC ...(((.((((((....((((((....))))))(((((((.((((.....))))))))))).............---.---------.))))-)).)))............. ( -20.30) >DroPse_CAF1 20556 111 + 1 UGUAGCAAGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUGCUUUUCUGCUUUAUUUUUCCCCAUUCUGUGCCCAC-UC ...((((((((((....((((((....))))))(((((((.((((.....))))))))))).....)))))))))).................................-.. ( -22.50) >DroSec_CAF1 20309 102 + 1 UGUAGCUGGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUUUCUU---------UUUUU-CCUGCUCUCUAACCGCCAC ...(((.((((((....((((((....))))))(((((((.((((.....))))))))))).................---------.))))-)).)))............. ( -20.30) >DroEre_CAF1 21132 99 + 1 UGUAGCUGGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUU---C---------UUUUU-CCUGCUCUCCGCCUGCUUU .((((.((((((.....((((((....))))))..............((((...((..(((((.....))))).---)---------)....-.)))))))))).))))... ( -25.50) >DroYak_CAF1 20048 95 + 1 UGUAGCUGGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUU---C---------UUUUU-CCUGCCCUCCGCCUG---- ..(((.(((((......((((((....))))))..............((((...((..(((((.....))))).---)---------)....-.)))).))))).)))---- ( -19.70) >DroPer_CAF1 20448 111 + 1 UGUAGCAAGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUGCUUUUCUGCUUUAUUUUUCCCCAUUCUGUGCCCAC-UC ...((((((((((....((((((....))))))(((((((.((((.....))))))))))).....)))))))))).................................-.. ( -22.50) >consensus UGUAGCUGGAGAACCAAUUUCCAACCAUGGAAACUUAAUUUAUGUCUGCAGCAUAGUUGAGAAUAUUUUUCUUU___C_________UUUUU_CCUGCUCUCUGCCCGC_UC ....((.(((((.....((((((....))))))(((((((.((((.....))))))))))).....................................)))))))....... (-15.61 = -16.00 + 0.39)

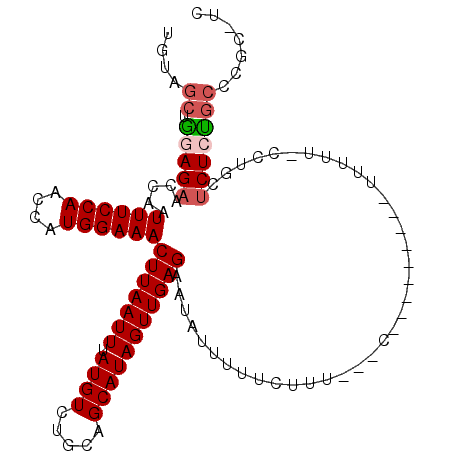
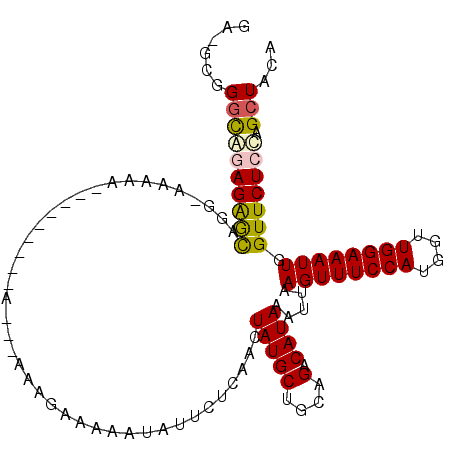
| Location | 17,412,962 – 17,413,061 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.65 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17412962 99 - 23771897 GUGGCGGGCAGAGAGCAGG-AAAAA---------G---AAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCCAGCUACA (((((.((.((((.((((.-(....---------.---..((((.....))))......).))))............((((((((....))))))))..)))))).))))). ( -26.42) >DroPse_CAF1 20556 111 - 1 GA-GUGGGCACAGAAUGGGGAAAAAUAAAGCAGAAAAGCAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCUUGCUACA ..-((((.((......((((((.......((((...((..((((.....))))...))...))))............((((((((....))))))))..)))))))))))). ( -23.10) >DroSec_CAF1 20309 102 - 1 GUGGCGGUUAGAGAGCAGG-AAAAA---------AAGAAAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCCAGCUACA (((((.....((((((((.-(....---------.((((.(......).))))......).))))............((((((((....))))))))...))))..))))). ( -23.60) >DroEre_CAF1 21132 99 - 1 AAAGCAGGCGGAGAGCAGG-AAAAA---------G---AAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCCAGCUACA ......((((((((((((.-(....---------.---..((((.....))))......).))))............((((((((....))))))))...))))).)))... ( -24.22) >DroYak_CAF1 20048 95 - 1 ----CAGGCGGAGGGCAGG-AAAAA---------G---AAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCCAGCUACA ----..((((((((((((.-(....---------.---..((((.....))))......).))))............((((((((....))))))))...))))).)))... ( -23.12) >DroPer_CAF1 20448 111 - 1 GA-GUGGGCACAGAAUGGGGAAAAAUAAAGCAGAAAAGCAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCUUGCUACA ..-((((.((......((((((.......((((...((..((((.....))))...))...))))............((((((((....))))))))..)))))))))))). ( -23.10) >consensus GA_GCGGGCAGAGAGCAGG_AAAAA_________A___AAAGAAAAAUAUUCUCAACUAUGCUGCAGACAUAAAUUAAGUUUCCAUGGUUGGAAAUUGGUUCUCCAGCUACA ......((((((((((.........................................(((((....).)))).....((((((((....)))))))).))))))).)))... (-17.76 = -17.65 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:41 2006