| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,955,758 – 1,955,872 |
| Length | 114 |
| Max. P | 0.785691 |

| Location | 1,955,758 – 1,955,872 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
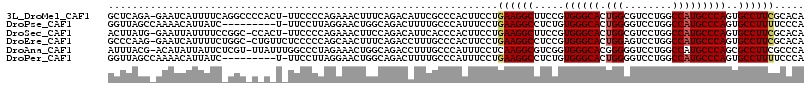
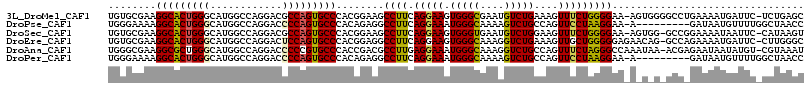
>3L_DroMel_CAF1 1955758 114 + 23771897 GCUCAGA-GAAUCAUUUUCAGGCCCCACU-UUCCCCAGAAACUUUCAGACAUUCGCCCACUUCCUGAAGGCUUCCGUGGGCACUGGCGUCCUGGCCAUGCCCAGUGCCUUCGCACA ((...((-((.....))))((((......-.......(((.(((((((...............))))))).)))..((((((.((((......))))))))))..))))..))... ( -32.76) >DroPse_CAF1 7101 106 + 1 GGUUAGCCAAAACAUUAUC---------U-UUCCUUAGGAACUGGCAGACUUUUGCCCAUUUCCUGAAGGCCUCUGUGGGCACUGGGGUCCUGGCCAUGCCCAGUGCCUUUUCCCA ((....))....(((....---------.-..(((((((((..(((((....)))))...)))))).))).....)))(((((((((((....))....)))))))))........ ( -34.90) >DroSec_CAF1 7072 113 + 1 ACUUAUG-GAAUUAUUUUCCGGC-CCACU-UUCCCCAGAAACUUCCAGACAUUCACCCACUUCCUGAAGGCUUCCGUGGGCACUGGCGUCCUGGCCAUGCCCAGUGCCUUCGCACA .....((-(((....((((.((.-.....-....)).)))).)))))..................((((((...(.((((((.((((......))))))))))).))))))..... ( -32.50) >DroEre_CAF1 6856 114 + 1 GCCCAAG-GAAUCAUUUUCUGGC-CUGUUCUCCCCCAGCAACUUUCAGACCUUUGCCCACUUCCUGAAGGCCUCCGUGGGCACUGGAGUCCUGGCCAUGCCCAGUGCCUUCGCACA (((..((-((.......(((((.-..(((((.....)).)))..)))))((..(((((((..((....)).....)))))))..))..)))))))........((((....)))). ( -33.20) >DroAna_CAF1 5619 114 + 1 AUUUACG-ACAUAUUAUUCUCGU-UUAUUUGGCCCUAGAAACUGGCAGACCUUUGCCCAUUUCCUCAAGGCGUCGGUGGGCACGGGGGUCCUGGCCAUGCCCAGCGCCUUCGCCCA .....((-((..........((.-.....))(((...((((..(((((....)))))..)))).....))))))).(((((..((((((.((((......)))).))))))))))) ( -36.90) >DroPer_CAF1 7804 106 + 1 GGUUAGCCAAAACAUUAUC---------U-UUCCUUAGGAACUGGCAGACUUUUGCCCAUUUCCUGAAGGCCUCUGUGGGCACUGGGGUCCUGGCCAUGCCCAGUGCCUUUUCCCA ((....))....(((....---------.-..(((((((((..(((((....)))))...)))))).))).....)))(((((((((((....))....)))))))))........ ( -34.90) >consensus GCUUAGG_AAAUCAUUUUC_GGC_C_A_U_UUCCCCAGAAACUGGCAGACAUUUGCCCACUUCCUGAAGGCCUCCGUGGGCACUGGGGUCCUGGCCAUGCCCAGUGCCUUCGCACA .................................................................((((((.....((((((.(((........)))))))))..))))))..... (-20.18 = -20.35 + 0.17)
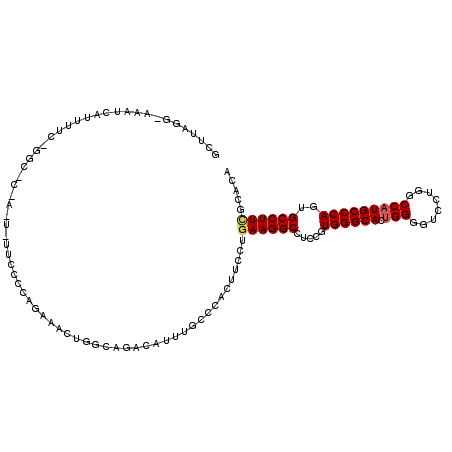
| Location | 1,955,758 – 1,955,872 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -38.89 |
| Consensus MFE | -22.61 |
| Energy contribution | -22.28 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
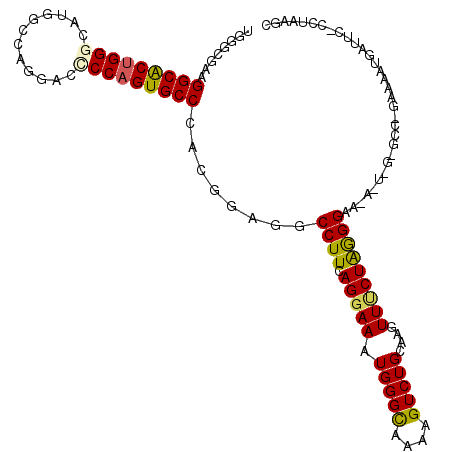
>3L_DroMel_CAF1 1955758 114 - 23771897 UGUGCGAAGGCACUGGGCAUGGCCAGGACGCCAGUGCCCACGGAAGCCUUCAGGAAGUGGGCGAAUGUCUGAAAGUUUCUGGGGAA-AGUGGGGCCUGAAAAUGAUUC-UCUGAGC .((((....)))).((((((...((((.(.(((.(.(((.(((((((.((((((..(....).....)))))).))))))))))..-).))).)))))...))).)))-....... ( -41.30) >DroPse_CAF1 7101 106 - 1 UGGGAAAAGGCACUGGGCAUGGCCAGGACCCCAGUGCCCACAGAGGCCUUCAGGAAAUGGGCAAAAGUCUGCCAGUUCCUAAGGAA-A---------GAUAAUGUUUUGGCUAACC ..((..((((((.(((((((((........))).))))))......((((.(((((...((((......))))..)))))))))..-.---------.....))))))..)).... ( -34.30) >DroSec_CAF1 7072 113 - 1 UGUGCGAAGGCACUGGGCAUGGCCAGGACGCCAGUGCCCACGGAAGCCUUCAGGAAGUGGGUGAAUGUCUGGAAGUUUCUGGGGAA-AGUGG-GCCGGAAAAUAAUUC-CAUAAGU .....((((((.(((((((((((......)))).)))))).)...)))))).....(((((......(((((.(.((((....)))-).)..-.))))).......))-))).... ( -41.32) >DroEre_CAF1 6856 114 - 1 UGUGCGAAGGCACUGGGCAUGGCCAGGACUCCAGUGCCCACGGAGGCCUUCAGGAAGUGGGCAAAGGUCUGAAAGUUGCUGGGGGAGAACAG-GCCAGAAAAUGAUUC-CUUGGGC .((((....)))).......(.((((((.((...(((((((.....((....))..)))))))..((((((....((.(....).))..)))-))).......)).))-).))).) ( -39.20) >DroAna_CAF1 5619 114 - 1 UGGGCGAAGGCGCUGGGCAUGGCCAGGACCCCCGUGCCCACCGACGCCUUGAGGAAAUGGGCAAAGGUCUGCCAGUUUCUAGGGCCAAAUAA-ACGAGAAUAAUAUGU-CGUAAAU ..(((.((((((((((((((((.........)))))))))..).))))))..((((((.((((......)))).))))))...)))......-((((.(......).)-))).... ( -42.90) >DroPer_CAF1 7804 106 - 1 UGGGAAAAGGCACUGGGCAUGGCCAGGACCCCAGUGCCCACAGAGGCCUUCAGGAAAUGGGCAAAAGUCUGCCAGUUCCUAAGGAA-A---------GAUAAUGUUUUGGCUAACC ..((..((((((.(((((((((........))).))))))......((((.(((((...((((......))))..)))))))))..-.---------.....))))))..)).... ( -34.30) >consensus UGGGCGAAGGCACUGGGCAUGGCCAGGACCCCAGUGCCCACGGAGGCCUUCAGGAAAUGGGCAAAAGUCUGCAAGUUUCUAGGGAA_A_U_G_GCC_GAAAAUGAUUC_CCUAAGC ........(((((((((............)))))))))........((((.(((((.(((((....)))))....)))))))))................................ (-22.61 = -22.28 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:47 2006