| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,374,949 – 17,375,042 |
| Length | 93 |
| Max. P | 0.827846 |

| Location | 17,374,949 – 17,375,042 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -21.83 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
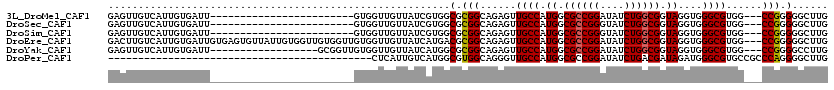
>3L_DroMel_CAF1 17374949 93 + 23771897 GAGUUGUCAUUGUGAUU------------------------GUGGUUGUUAUCGUGGCGCGGCAGAGUUGCCAUGGCGCCGGAUAUCUGGCGGUAGGUGGGCGUGG---CCGGGGGCUUG (((((.((...(((.(.------------------------.((((....))))..))))(((...((..((.((.((((((....)))))).))))..).)...)---)))).))))). ( -36.40) >DroSec_CAF1 1860 93 + 1 GAGUUGUCAUUGUGAUU------------------------GUGGUUGUUAUCGUGGCGCGGCAGAGUUGCCAUGGCGCCGGGUAUCUGGCGGUAGGUGGGCGUGG---CCGGGGGCUUG (((((.((...(((.(.------------------------.((((....))))..))))(((...((..((.((.((((((....)))))).))))..).)...)---)))).))))). ( -35.50) >DroSim_CAF1 1559 93 + 1 GAGUUGUCAUUGUGAUU------------------------GUGGUUGUUAUCGUGGCGCGGCAGAGUUGCCAUGGCGCCGGGUAUCUGGCGGUAGGUGGGCGUGG---CCGGGGGCUUG (((((.((...(((.(.------------------------.((((....))))..))))(((...((..((.((.((((((....)))))).))))..).)...)---)))).))))). ( -35.50) >DroEre_CAF1 1901 117 + 1 GACUUGUCAUUGUGAUUGUGAGUGUUAUUGUGGUUGUGGUUGUGGUUGUUAUCAUGACGCGGCAGAGUUGCCAUGGCGCCGGAUAUCUGGCGGUAGGUGGGCGUGG---CCGGGGGCUUG .....(((...(..((..(((......)))..))..).((..((((....))))..)).((((...((..((.((.((((((....)))))).))))..).)...)---)))..)))... ( -38.10) >DroYak_CAF1 2027 99 + 1 GAGUUGUCAUUGUGAUU------------------GCGGUUGUGGUUGUUAUCAUGGCGCGGCAGAGUUGCCAUGGCGCCGGAUAUCUGGCGGUAGGUGGGCGUGG---CCGGGGCCUUG ..((.(((.....))).------------------))((((.((((..(..(((((((((......).))))))))((((((....))))))..........)..)---))).))))... ( -37.20) >DroPer_CAF1 467 76 + 1 --------------------------------------------CUCAUUGUCAUGGCGUGGCAGGGUUGCCAUGGCGCCGGAUAUCUGACGAUAGAUGGGCGUGCCGCCCAGGGGCUUG --------------------------------------------......(((((((((.........)))))))))(((...((((....))))..((((((...))))))..)))... ( -29.70) >consensus GAGUUGUCAUUGUGAUU________________________GUGGUUGUUAUCAUGGCGCGGCAGAGUUGCCAUGGCGCCGGAUAUCUGGCGGUAGGUGGGCGUGG___CCGGGGGCUUG .........................................................(.(((......((((.((.((((((....)))))).))....))))......))).)...... (-21.83 = -21.72 + -0.11)

| Location | 17,374,949 – 17,375,042 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -14.03 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
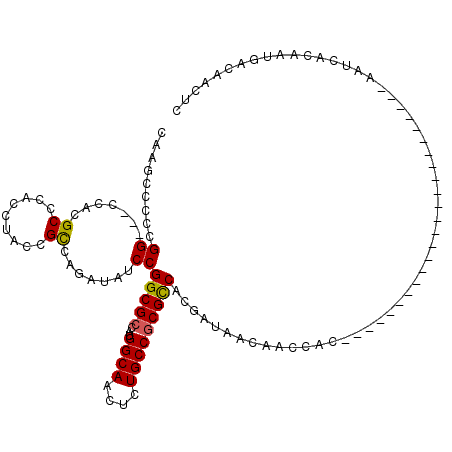
>3L_DroMel_CAF1 17374949 93 - 23771897 CAAGCCCCCGG---CCACGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAACAACCAC------------------------AAUCACAAUGACAACUC .........((---(..............))).(.((((.(((((...((((....)))))))))..)))).)......------------------------................. ( -19.14) >DroSec_CAF1 1860 93 - 1 CAAGCCCCCGG---CCACGCCCACCUACCGCCAGAUACCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAACAACCAC------------------------AAUCACAAUGACAACUC .........((---(..............)))........(((((...((((....)))))))))..............------------------------................. ( -17.74) >DroSim_CAF1 1559 93 - 1 CAAGCCCCCGG---CCACGCCCACCUACCGCCAGAUACCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAACAACCAC------------------------AAUCACAAUGACAACUC .........((---(..............)))........(((((...((((....)))))))))..............------------------------................. ( -17.74) >DroEre_CAF1 1901 117 - 1 CAAGCCCCCGG---CCACGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGUCAUGAUAACAACCACAACCACAACCACAAUAACACUCACAAUCACAAUGACAAGUC ...(((...))---)...((........((((.(....).))))....((((....))))))(((((...........................................)))))..... ( -18.33) >DroYak_CAF1 2027 99 - 1 CAAGGCCCCGG---CCACGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGCCAUGAUAACAACCACAACCGC------------------AAUCACAAUGACAACUC ...(((....)---))..((....((......)).((((.(((((...((((....)))))))))..))))............))------------------................. ( -22.40) >DroPer_CAF1 467 76 - 1 CAAGCCCCUGGGCGGCACGCCCAUCUAUCGUCAGAUAUCCGGCGCCAUGGCAACCCUGCCACGCCAUGACAAUGAG-------------------------------------------- ...(((....)))(((.((((....((((....))))...))))...(((((....))))).)))...........-------------------------------------------- ( -26.10) >consensus CAAGCCCCCGG___CCACGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAACAACCAC________________________AAUCACAAUGACAACUC .........((.......((.........)).......))(((((...((((....)))))))))....................................................... (-14.03 = -13.92 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:24 2006