
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,354,131 – 17,354,230 |
| Length | 99 |
| Max. P | 0.951910 |

| Location | 17,354,131 – 17,354,230 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17354131 99 + 23771897 AC-CGAUCGUAUCAA-AUUCAAUGGACG-AUUGAAUU--UGAUCGAUGCCAA-GC-----U---AAGUUGGUUU-----AACUUCUAGGUCGAGUGUGGAAUACCAACUAAAGCAUCA ..-..((((.(((((-(((((((.....-))))))))--)))))))).....-((-----(---.(((((((..-----...(((((.(.....).))))).)))))))..))).... ( -29.30) >DroVir_CAF1 43200 106 + 1 GUACGAUCGUAUCAAAAUUCAAUGGUCA-AUUGAAUUUUUGAUCGAUGCCAA-GC-----AAAAAAGUUGCUUC-----AACUUCUGGGUCGACACUGGGCUAGCAACUAAAGCAUCA ....(((((....((((((((((.....-)))))))))))))))(((((.((-((-----((.....)))))).-----.....((((.(((....))).))))........))))). ( -27.60) >DroGri_CAF1 34053 93 + 1 UC-CGAUCGUAUCAAAAUUCAAUGGUCA-AUUGAAUUUUUGAUCGAUGCCAA-GC-----AGAAAAGUUGCCUC-----AA------------CACUGGGCUAGCAACUAAAGCAUCA ..-.(((((....((((((((((.....-)))))))))))))))((((((..-..-----.)...(((((((((-----(.------------...))))...))))))...))))). ( -25.00) >DroWil_CAF1 40996 112 + 1 AC-CGAUCGUAUCAAAAUUCAAUGGUCAAAUUGAAUU--UGAUCGAUGCCCAAACUUUUGU---UGGUUGGUUUAUUUAAACUUCUAGGUCGAGUGUGGGAUACCAACUAAAGCAUCA ..-.(((((......((((((((......))))))))--)))))(((((.(((....)))(---((((((((..((((..((((.......))))...))))))))))))).))))). ( -27.90) >DroMoj_CAF1 48053 105 + 1 GU-CGAUCGUAUCAAAAUUCAAUGGUCA-AUUGAAUUUUGGAUCGAUGCUGA-GC-----AAAAAAAUUGCCUC-----AACUUCUGGGUCGACUCUGGGCUAGCAACUAAAGCAUCA ..-.((((....(((((((((((.....-)))))))))))))))((((((((-((-----((.....))).)))-----.....((((.(((....))).)))).......)))))). ( -30.40) >DroPer_CAF1 37447 100 + 1 AC-CGAUCGUAUCAAAAUUCAAUGGUCA-AUUGAAUU--UGAUCGAUGCCCA-GC-----U---AAGUUGGUUU-----AACUUCUAGGUCGAGUGUGGAAUACCAACUAAAGCAUCA ..-..((((.((((((.((((((.....-))))))))--)))))))).....-((-----(---.(((((((..-----...(((((.(.....).))))).)))))))..))).... ( -26.20) >consensus AC_CGAUCGUAUCAAAAUUCAAUGGUCA_AUUGAAUU__UGAUCGAUGCCAA_GC_____A___AAGUUGCUUC_____AACUUCUAGGUCGACUCUGGGAUACCAACUAAAGCAUCA ....(((((......((((((((......))))))))..)))))(((((................(((((((..............................)))))))...))))). (-16.46 = -16.65 + 0.19)

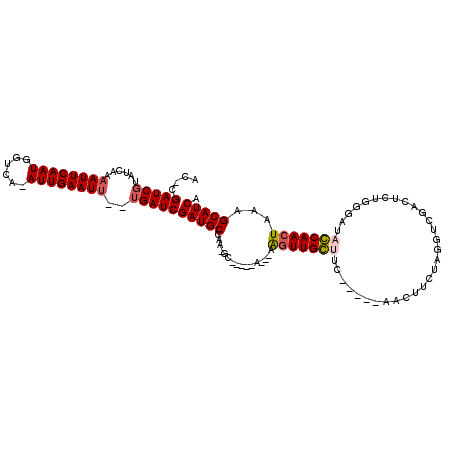
| Location | 17,354,131 – 17,354,230 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -18.94 |
| Energy contribution | -18.36 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17354131 99 - 23771897 UGAUGCUUUAGUUGGUAUUCCACACUCGACCUAGAAGUU-----AAACCAACUU---A-----GC-UUGGCAUCGAUCA--AAUUCAAU-CGUCCAUUGAAU-UUGAUACGAUCG-GU .((((((..(((((((.......((((......).))).-----..))))))).---.-----..-..)))))).((((--((((((((-.....)))))))-))))).......-.. ( -26.10) >DroVir_CAF1 43200 106 - 1 UGAUGCUUUAGUUGCUAGCCCAGUGUCGACCCAGAAGUU-----GAAGCAACUUUUUU-----GC-UUGGCAUCGAUCAAAAAUUCAAU-UGACCAUUGAAUUUUGAUACGAUCGUAC .((((((..(((((((.........(((((......)))-----))))))))).....-----..-..))))))((((.((((((((((-.....)))))))))).....)))).... ( -27.10) >DroGri_CAF1 34053 93 - 1 UGAUGCUUUAGUUGCUAGCCCAGUG------------UU-----GAGGCAACUUUUCU-----GC-UUGGCAUCGAUCAAAAAUUCAAU-UGACCAUUGAAUUUUGAUACGAUCG-GA ....((.......))..(((.((((------------..-----(((....)))..).-----))-).))).((((((.((((((((((-.....)))))))))).....)))))-). ( -25.30) >DroWil_CAF1 40996 112 - 1 UGAUGCUUUAGUUGGUAUCCCACACUCGACCUAGAAGUUUAAAUAAACCAACCA---ACAAAAGUUUGGGCAUCGAUCA--AAUUCAAUUUGACCAUUGAAUUUUGAUACGAUCG-GU ....(((((.((((((.........((......)).((((....))))..))))---)).)))))....(.((((((((--((((((((......)))))).)))))).))))).-.. ( -22.80) >DroMoj_CAF1 48053 105 - 1 UGAUGCUUUAGUUGCUAGCCCAGAGUCGACCCAGAAGUU-----GAGGCAAUUUUUUU-----GC-UCAGCAUCGAUCCAAAAUUCAAU-UGACCAUUGAAUUUUGAUACGAUCG-AC .((((((..(((.....((((((..((......))..))-----).))).........-----))-).))))))(((((((((((((((-.....)))))))))))....)))).-.. ( -28.74) >DroPer_CAF1 37447 100 - 1 UGAUGCUUUAGUUGGUAUUCCACACUCGACCUAGAAGUU-----AAACCAACUU---A-----GC-UGGGCAUCGAUCA--AAUUCAAU-UGACCAUUGAAUUUUGAUACGAUCG-GU .(((((((.(((((((.......((((......).))).-----..))))))).---.-----..-.)))))))(((((--((((((((-.....)))))))))......)))).-.. ( -23.70) >consensus UGAUGCUUUAGUUGCUAGCCCACACUCGACCUAGAAGUU_____AAACCAACUU___A_____GC_UUGGCAUCGAUCA__AAUUCAAU_UGACCAUUGAAUUUUGAUACGAUCG_GU .((((((..(((((((.........((......))...........)))))))...............))))))((((...((((((((......)))))))).......)))).... (-18.94 = -18.36 + -0.58)

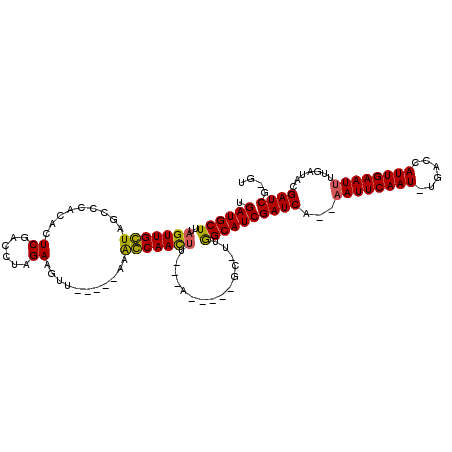
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:20 2006