
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,331,010 – 17,331,107 |
| Length | 97 |
| Max. P | 0.973833 |

| Location | 17,331,010 – 17,331,107 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.37 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
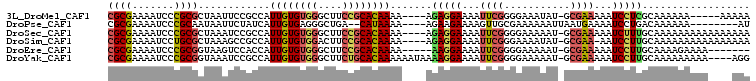
>3L_DroMel_CAF1 17331010 97 + 23771897 CGCGAAAAUCCCGCGCUAAUUCCGCCAUUGUGUGGGCUUCCGCACAAAA----AGAGGAAAAUUCGGGGAAAUAU-GCGAAAAAUCCUCGCAAAAAA-----AAAAA .((((........(((((.((((.((.((((((((....))))))))..----.(((.....))))))))).)).-)))........))))......-----..... ( -28.99) >DroPse_CAF1 15523 93 + 1 CGCGAAAAUCCCGCAAUAAUUCUAUCAUUGUGAGGGCUGA--CAUAAAA----AGAAGAAAAGUUGCGAAAAAAUUAAUGAAAAUCCUGACAAAAAA--------AU ...........((((((..((((.((.(((((........--)))))..----.))))))..)))))).............................--------.. ( -11.20) >DroSec_CAF1 12764 102 + 1 CGCGAAAAUCCCGCGCUAAAUCCGCCAUUGUGUGGGCUUCCGCACAAAA----AGAGGAAAAUUCGGGGAAAAAU-GCGAAAAAUCUUUGCAAAAAAAAAAAAAAAA .(((((.......(((....(((.((.((((((((....))))))))..----.(((.....)))))))).....-))).......)))))................ ( -26.24) >DroSim_CAF1 12633 101 + 1 CGCGAAAAUCCUGCGCUAAAGCCGCCAUUGUGUGGACUUCCGCACAAAA----AGAGGAAAAUUCGGGAAAAUAU-GCGAA-AAUCCUUGCAAAAAAAAAAAAAAAA (.((((..((((((((....).)))..((((((((....))))))))..----..))))...)))).)......(-((((.-.....)))))............... ( -26.40) >DroEre_CAF1 19294 94 + 1 CGCGAAAAUCCCGCGGUAAGUCCACCAUUGUGUGGGCUUCCGCACAAAA-----AAGGAAAAUUCGGGGAAAAAU-GCGAAAAAUCCUUGCAAAAGAAAA------- .((.........((((.((((((((......))))))))))))......-----(((((...((((.(......)-.))))...))))))).........------- ( -30.20) >DroYak_CAF1 19428 102 + 1 CGCGAAAAUCCCGCGGUAAAUCCGCCAUUGUGUGGGCUUCUGCACAAAAAAUAAAAGGAAAAUUCGGGGAAAAAU-GCGAAAAAUCCUUGCAAAAAAAAA----AGG ((((....((((((((.....))))..(((((..(....)..)))))...................))))....)-)))......((((..........)----))) ( -26.00) >consensus CGCGAAAAUCCCGCGCUAAAUCCGCCAUUGUGUGGGCUUCCGCACAAAA____AGAGGAAAAUUCGGGGAAAAAU_GCGAAAAAUCCUUGCAAAAAAAAA____AAA ((((.......))))............((((((((....)))))))).......(((((...((((...........))))...))))).................. (-15.78 = -16.37 + 0.59)

| Location | 17,331,010 – 17,331,107 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -12.89 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
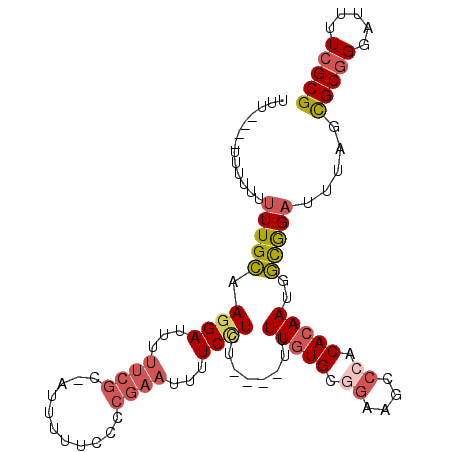
>3L_DroMel_CAF1 17331010 97 - 23771897 UUUUU-----UUUUUUGCGAGGAUUUUUCGC-AUAUUUCCCCGAAUUUUCCUCU----UUUUGUGCGGAAGCCCACACAAUGGCGGAAUUAGCGCGGGAUUUUCGCG .....-----......((((((((((..(((-.((.((((((((.......)).----..(((((.((....)).))))).)).)))).)))))..)))))))))). ( -31.50) >DroPse_CAF1 15523 93 - 1 AU--------UUUUUUGUCAGGAUUUUCAUUAAUUUUUUCGCAACUUUUCUUCU----UUUUAUG--UCAGCCCUCACAAUGAUAGAAUUAUUGCGGGAUUUUCGCG ..--------..........................(..(((((...((((((.----..((.((--........)).)).)).))))...)))))..)........ ( -10.00) >DroSec_CAF1 12764 102 - 1 UUUUUUUUUUUUUUUUGCAAAGAUUUUUCGC-AUUUUUCCCCGAAUUUUCCUCU----UUUUGUGCGGAAGCCCACACAAUGGCGGAUUUAGCGCGGGAUUUUCGCG ............................(((-.....((((((.....(((.((----..(((((.((....)).))))).)).))).....)).)))).....))) ( -24.90) >DroSim_CAF1 12633 101 - 1 UUUUUUUUUUUUUUUUGCAAGGAUU-UUCGC-AUAUUUUCCCGAAUUUUCCUCU----UUUUGUGCGGAAGUCCACACAAUGGCGGCUUUAGCGCAGGAUUUUCGCG ............((((((..(((((-(((((-(((.......((....))....----...)))))))))))))........((.......))))))))........ ( -24.84) >DroEre_CAF1 19294 94 - 1 -------UUUUCUUUUGCAAGGAUUUUUCGC-AUUUUUCCCCGAAUUUUCCUU-----UUUUGUGCGGAAGCCCACACAAUGGUGGACUUACCGCGGGAUUUUCGCG -------.........(((((((...((((.-.........))))...)))))-----..((.((((((((.((((......)))).))).))))).)).....)). ( -26.30) >DroYak_CAF1 19428 102 - 1 CCU----UUUUUUUUUGCAAGGAUUUUUCGC-AUUUUUCCCCGAAUUUUCCUUUUAUUUUUUGUGCAGAAGCCCACACAAUGGCGGAUUUACCGCGGGAUUUUCGCG ...----...........(((((...((((.-.........))))...))))).........(((.((((.(((.(.....)((((.....))))))).))))))). ( -20.70) >consensus UUU____UUUUUUUUUGCAAGGAUUUUUCGC_AUUUUUCCCCGAAUUUUCCUCU____UUUUGUGCGGAAGCCCACACAAUGGCGGAUUUAGCGCGGGAUUUUCGCG .............(((((.((((...((((...........))))...))))........(((((.((....)).)))))..))))).....(((((.....))))) (-12.89 = -13.83 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:17 2006