
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,309,705 – 17,309,859 |
| Length | 154 |
| Max. P | 0.694079 |

| Location | 17,309,705 – 17,309,819 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -27.28 |
| Energy contribution | -26.65 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17309705 114 + 23771897 GCCAGGUGGCAUAAUCGAUGCUCUGAAUGAAGAAACGAAUGGGCUCCAGGAUAAGAUAUUGGAAAAAGAAGACCAGGACGAUGGUUACCAACAUCGUCUUGAAUCUCUGGGGCU .(((..((((((.....))))(((......)))..))..)))((((((((((......((....)).......((((((((((........)))))))))).))).))))))). ( -37.90) >DroPse_CAF1 7632 114 + 1 GCCAGGUGGCCGAGUCGAUGGACAGCACAAUGAAUCUGAUAGGAUCGAGGACGAUGAACUGGAACAGCAAUACUAGCACAAUCGUCACCACCAUGGUCUUGAACUUCUUGGUCU ((((((.(..((((.(.((((..........((.(((....)))))(..((((((...((((..........))))....))))))..).)))).).))))..)..)))))).. ( -31.90) >DroSec_CAF1 8622 114 + 1 GCCAGGUGGCAUAAUCGAUGCUCAGAAUGAAGAAACGAAUGGGCUCCAGGAUGAGGUAUUGGAAAAAGAAGACCAGGACGAUGGUUACCAACAUCGUCUUGAGUCUCUGGGGCU .(((..((((((.....))))((.....)).....))..)))(((((((.........((....))...((((((((((((((........)))))))))).))))))))))). ( -38.60) >DroSim_CAF1 7517 114 + 1 GCCAGGUGGCAUAAUCGAUGCUCAGAAUGAAGAAACGAAUGGGCUCCAGGAUGAGGUAUUGGAAAAAGAAGACCAGGACGAUGGUUACCAACAUCGUCUUGAGUCUCUGGGGCU .(((..((((((.....))))((.....)).....))..)))(((((((.........((....))...((((((((((((((........)))))))))).))))))))))). ( -38.60) >DroEre_CAF1 6929 114 + 1 GCCAGGUGGCAUAGUCGAUGCCCAGAACGAAGAAACGAAAGGGCUCUAGGACGAGAUAUUGGAAAAAGAACACCAGGACGAUGGUCACCAACAUCGUCUUGAAUCUCUGCGGCU (((....)))..(((((..((((....((......))...))))........(((((.((....)).......((((((((((........)))))))))).)))))..))))) ( -35.90) >DroYak_CAF1 8963 114 + 1 GCCAGGUGGCAUAAUCGAUGCCCAGAAUGAGGAAACGGAUGGGUUCUAGAAUGAGAUAUUGGAAAAAGAACACCAGGACGAUGGUCACCAACAUCGUCUUGAAUCUCUGGGGCU (((....)))..........((((((....(....).(((..(((((...................)))))..((((((((((........)))))))))).)))))))))... ( -35.61) >consensus GCCAGGUGGCAUAAUCGAUGCUCAGAAUGAAGAAACGAAUGGGCUCCAGGAUGAGAUAUUGGAAAAAGAAGACCAGGACGAUGGUCACCAACAUCGUCUUGAAUCUCUGGGGCU (((....)))...............................(((((((((.(((....)))............((((((((((........))))))))))....))))))))) (-27.28 = -26.65 + -0.63)

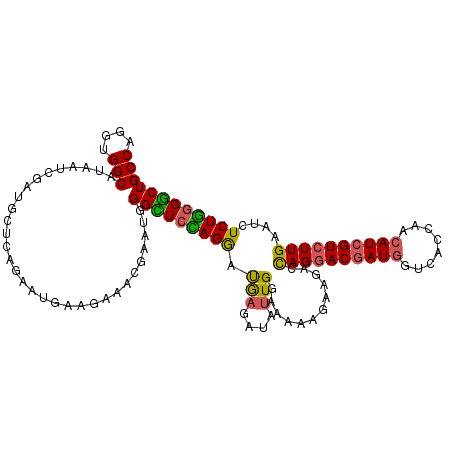

| Location | 17,309,745 – 17,309,859 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17309745 114 + 23771897 GGGCUCCAGGAUAAGAUAUUGGAAAAAGAAGACCAGGACGAUGGUUACCAACAUCGUCUUGAAUCUCUGGGGCUCGUGCAUGAAUCCCGAGAAGAUUGAGACCAGAACGAAGCA ((((((((((((......((....)).......((((((((((........)))))))))).))).)))))))))((...((..((.(((.....))).)).))..))...... ( -35.10) >DroPse_CAF1 7672 114 + 1 AGGAUCGAGGACGAUGAACUGGAACAGCAAUACUAGCACAAUCGUCACCACCAUGGUCUUGAACUUCUUGGUCUCGUGUUGGAAGCCUGUGAUCACGGCUAUUAGGGUGAAACU ..(((((((((((((...((((..........))))....))))))(((.....))).........)))))))(((..((((.((((.((....)))))).))))..))).... ( -28.50) >DroSec_CAF1 8662 114 + 1 GGGCUCCAGGAUGAGGUAUUGGAAAAAGAAGACCAGGACGAUGGUUACCAACAUCGUCUUGAGUCUCUGGGGCUCGUGCAUAAAUCCCGAGAAGAUUGCGACCAGGAUGAAGCA (((((((((.........((....))...((((((((((((((........)))))))))).))))))))))))).(((....((((.(.(..(....)..)).))))...))) ( -40.40) >DroSim_CAF1 7557 114 + 1 GGGCUCCAGGAUGAGGUAUUGGAAAAAGAAGACCAGGACGAUGGUUACCAACAUCGUCUUGAGUCUCUGGGGCUCGUGCAUAAAUCCCGAAAAGAUUGCGACCAGGAUGAAGCA (((((((((.........((....))...((((((((((((((........)))))))))).))))))))))))).(((....((((.(.............).))))...))) ( -39.72) >DroEre_CAF1 6969 114 + 1 GGGCUCUAGGACGAGAUAUUGGAAAAAGAACACCAGGACGAUGGUCACCAACAUCGUCUUGAAUCUCUGCGGCUCGUGCAUGAAUCCAGAGAACAUUGAUACCAGGAUGAAGCA ((.(((......))).(((..(...........((((((((((........))))))))))..((((((.(..(((....)))..)))))))...)..)))))........... ( -31.70) >DroYak_CAF1 9003 114 + 1 GGGUUCUAGAAUGAGAUAUUGGAAAAAGAACACCAGGACGAUGGUCACCAACAUCGUCUUGAAUCUCUGGGGCUCGUGUAUGAAUCCCGAGAAGAUUGAUACCAGGGUGAAGCA ((((((((((........((....)).......((((((((((........))))))))))....))))))))))(.((((.((((.......)))).)))))........... ( -33.60) >consensus GGGCUCCAGGAUGAGAUAUUGGAAAAAGAAGACCAGGACGAUGGUCACCAACAUCGUCUUGAAUCUCUGGGGCUCGUGCAUGAAUCCCGAGAAGAUUGAGACCAGGAUGAAGCA ((((((((((.(((....)))............((((((((((........))))))))))....))))))))))..((....((((.(.............).))))...)). (-26.58 = -26.00 + -0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:13 2006