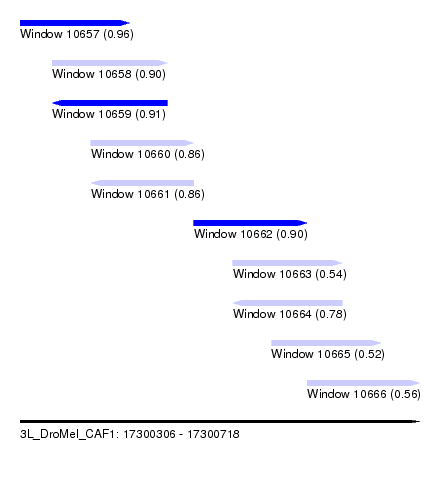
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,300,306 – 17,300,718 |
| Length | 412 |
| Max. P | 0.959463 |

| Location | 17,300,306 – 17,300,419 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.68 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300306 113 + 23771897 GCAUAUUCCUGCUCCACAUCCCCGUUUGG-------CAAAAGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAA ((...((((((((..((......))..))-------)...))))).........(((.....((.((((((((........)))))))).)).(((((((....)))))))))))).... ( -25.20) >DroSim_CAF1 807 111 + 1 GCAUAUUCCUGCUCCACAUCCUC--UUGG-------AAAAAGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAA ((...(((((..((((.......--.)))-------)...))))).........(((.....((.((((((((........)))))))).)).(((((((....)))))))))))).... ( -27.40) >DroEre_CAF1 4509 112 + 1 GCAUAUUCCUGCGCCACAUCCCC--UUGG------AAAAAAGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAA .........(((((((.....((--((..------....))))..........(((((....((.((((((((........)))))))).)).....)))))........)))))))... ( -27.00) >DroYak_CAF1 4829 118 + 1 GCAUAUUCCUGCUCCACAUCCCC--UUGGCAAGAAAAAAAAGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAA ((...(((((.(((((.......--.)))..)).......))))).........(((.....((.((((((((........)))))))).)).(((((((....)))))))))))).... ( -22.60) >consensus GCAUAUUCCUGCUCCACAUCCCC__UUGG_______AAAAAGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAA ((...(((((.........((......))...........))))).........(((.....((.((((((((........)))))))).)).(((((((....)))))))))))).... (-22.75 = -22.75 + 0.00)

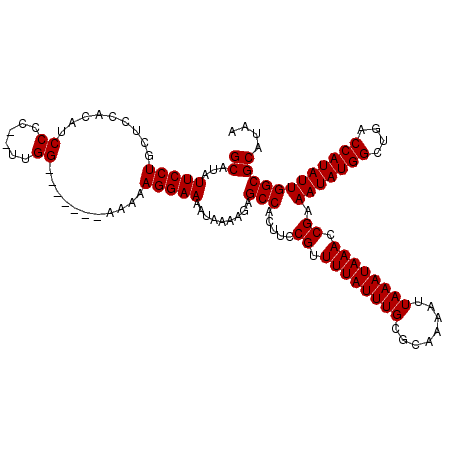
| Location | 17,300,339 – 17,300,458 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.31 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300339 119 + 23771897 AGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCCCA- .((...........(((.....((.((((((((........)))))))).)).(((((((....))))))))))((((...(((((...................))))).)))).)).- ( -27.31) >DroSim_CAF1 838 120 + 1 AGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCCCAC .((...........(((.....((.((((((((........)))))))).)).(((((((....))))))))))((((...(((((...................))))).)))).)).. ( -27.31) >DroEre_CAF1 4541 120 + 1 AGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCUCAU ............(((((.....((.((((((((........)))))))).)).(((((((....))))))))))((((...(((((...................))))).)))).)).. ( -25.41) >DroYak_CAF1 4867 120 + 1 AGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCACAU ..............(((.....((.((((((((........)))))))).)).(((((((....))))))))))((((...(((((...................))))).))))..... ( -25.31) >consensus AGGAAAAUAAAAGAGCCACUUCCGUUUUAUUUGCGCAAAAUUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCCCAU ..............(((.....((.((((((((........)))))))).)).(((((((....))))))))))((((...(((((...................))))).))))..... (-25.31 = -25.31 + 0.00)

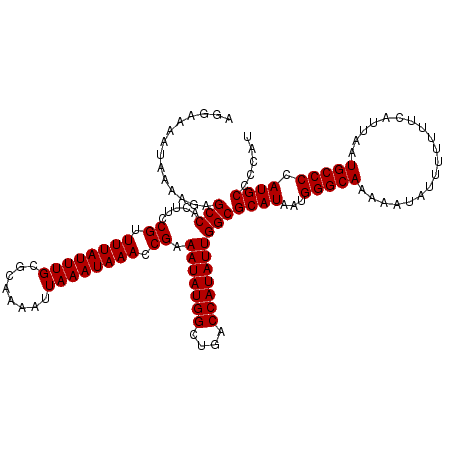

| Location | 17,300,339 – 17,300,458 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300339 119 - 23771897 -UGGGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAAUUUUGCGCAAAUAAAACGGAAGUGGCUCUUUUAUUUUCCU -(((.((((((((((..((((.......))))..)))))..))))).)))........((((((.(((((.(((((((..........))))))).)))))))))))............. ( -33.70) >DroSim_CAF1 838 120 - 1 GUGGGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAAUUUUGCGCAAAUAAAACGGAAGUGGCUCUUUUAUUUUCCU .(((.((((((((((..((((.......))))..)))))..))))).)))........((((((.(((((.(((((((..........))))))).)))))))))))............. ( -33.90) >DroEre_CAF1 4541 120 - 1 AUGAGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAAUUUUGCGCAAAUAAAACGGAAGUGGCUCUUUUAUUUUCCU .....((((((((((..((((.......))))..)))))..)))))((((.(((((....)))))(((((.(((((((..........))))))).))))).)))).............. ( -30.60) >DroYak_CAF1 4867 120 - 1 AUGUGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAAUUUUGCGCAAAUAAAACGGAAGUGGCUCUUUUAUUUUCCU .....((((((((((..((((.......))))..)))))..)))))((((.(((((....)))))(((((.(((((((..........))))))).))))).)))).............. ( -30.60) >consensus AUGGGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAAUUUUGCGCAAAUAAAACGGAAGUGGCUCUUUUAUUUUCCU .....((((((((((..((((.......))))..)))))..)))))((((.(((((....)))))(((((.(((((((..........))))))).))))).)))).............. (-30.60 = -30.60 + -0.00)


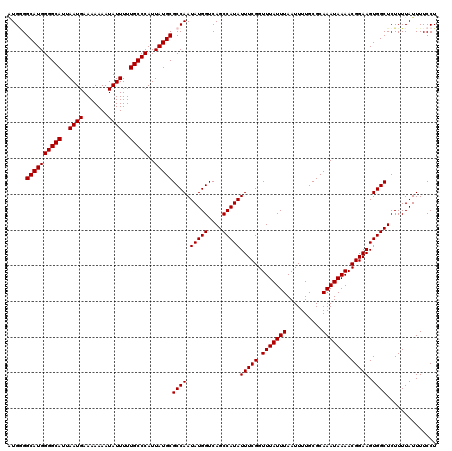
| Location | 17,300,379 – 17,300,485 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.37 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300379 106 + 23771897 UUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCCCA--------------UACCCCAGCGGAGUAGAUGGCAAAAUA .............(((((((....)))))))((.((((((((((.((((....)))))))))).)))).)).((((...--------------(((((....)).)))...))))..... ( -24.80) >DroSim_CAF1 878 120 + 1 UUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCCCACGCCCCAUACCCCGUGCCCCAGCGGAGUAGAUGGCAAAAUA .............(((((((....)))))))((.((((...(((((...................))))).)))).))..(((...(((.((((......)))).)))...)))...... ( -32.61) >DroEre_CAF1 4581 106 + 1 UUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCUCAUGCC--------------CCAGCGGAGUAGAUGGCAAAAUA .............(((((((....)))))))((.((((((((((.((((....)))))))))).)))).)).((((...((((--------------(....)).)))...))))..... ( -25.50) >DroYak_CAF1 4907 113 + 1 UUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCACAUGCCCCA-------UUCCCCAGCGGAGUAGAUGGCAAAAUA .............(((((((....)))))))((.((((((((((.((((....)))))))))).)))).)).(((((..((((((.-------.......).)).)))..)))))..... ( -27.30) >consensus UUAAAUAAACCGAAAUAUGGCUGACCAUAUUGGCGCAUAAUGGGCAAAAAUAUUUUUUUCAUUAAUGCCCCAUGCCCCAUGCC__________U_CCCCAGCGGAGUAGAUGGCAAAAUA .........(((.(((((((....)))))))((.((((...(((((...................))))).)))).)).......................)))................ (-21.01 = -21.14 + 0.12)

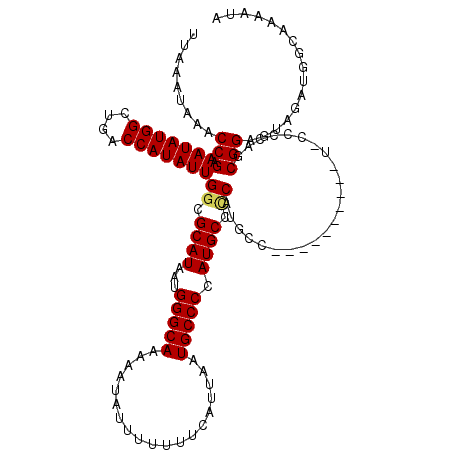

| Location | 17,300,379 – 17,300,485 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.37 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300379 106 - 23771897 UAUUUUGCCAUCUACUCCGCUGGGGUA--------------UGGGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAA ......(((...((((((...))))))--------------(((.((((((((((..((((.......))))..)))))..))))).)))((((((....))))))...)))........ ( -33.10) >DroSim_CAF1 878 120 - 1 UAUUUUGCCAUCUACUCCGCUGGGGCACGGGGUAUGGGGCGUGGGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAA ......(((...(((((((........)))))))...))).(((.((((((((((..((((.......))))..)))))..))))).)))((((((....)))))).............. ( -40.60) >DroEre_CAF1 4581 106 - 1 UAUUUUGCCAUCUACUCCGCUGG--------------GGCAUGAGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAA ......(((............((--------------.((((((......(((((..((((.......))))..))))).)))))).))(((((((....)))))))..)))........ ( -29.90) >DroYak_CAF1 4907 113 - 1 UAUUUUGCCAUCUACUCCGCUGGGGAA-------UGGGGCAUGUGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAA .....((((..(((.(((.....))).-------)))))))....((((((((((..((((.......))))..)))))..)))))((((((((((....)))))))..)))........ ( -32.40) >consensus UAUUUUGCCAUCUACUCCGCUGGGG_A__________GGCAUGGGGCAUGGGGCAUUAAUGAAAAAAAUAUUUUUGCCCAUUAUGCGCCAAUAUGGUCAGCCAUAUUUCGGUUUAUUUAA ..................((((((.................(((.((((((((((..((((.......))))..)))))..))))).)))((((((....))))))))))))........ (-25.12 = -25.88 + 0.75)

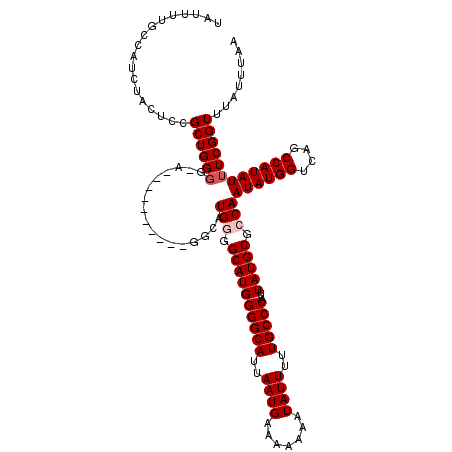
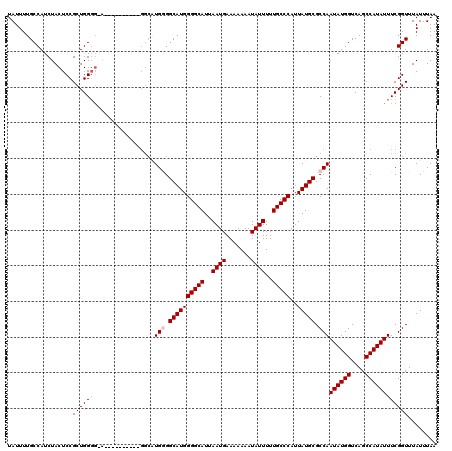
| Location | 17,300,485 – 17,300,602 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.78 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -28.10 |
| Energy contribution | -28.61 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300485 117 + 23771897 AAUAAAUAAAUAAGCGGAGCCAAAAAGGAAAGCCGAAAAGUAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGAAUCCA---GUUUGGUGGCUA ............((((((((......((....))...........)))))).)).(((((((..(((((.....))))).)))))))....(((((.(((((...---))))).))))). ( -30.13) >DroSim_CAF1 998 117 + 1 AAUAAAUAAAUAAGCGGAGCCAAAAAGGAAAGCCGAAAAGUAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCA---GUUUGGUGGCUA ............(((...((((((..(((...(((...(((....)))....((((((((((..(((((.....))))).)))))))..))).....))).))).---.)))))).))). ( -30.70) >DroEre_CAF1 4687 112 + 1 AA----UAAAUAAGCGGAGCCAAAAAGGAAAGCCGAAAAGUAAAAGCCCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCA----GUUGGUGGCUA ..----........(((..((.....))....))).........((((((((((((((((((..(((((.....))))).))))))).((........))...))----))))).)))). ( -34.20) >DroYak_CAF1 5020 116 + 1 AA----UAAAUAAGCGGAGCCAAAAAGGAAAGCCGAAAAGUAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCACCAAACUGGUGGCUA ..----......((((((((......((....))...........)))))).))((.(((((..(((((.....)))))((((((.((((........)).)).))))))))))).)).. ( -32.13) >consensus AA____UAAAUAAGCGGAGCCAAAAAGGAAAGCCGAAAAGUAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCA___GUUUGGUGGCUA ............((((((((......((....))...........)))))).)).(((((((..(((((.....))))).)))))))....(((((.(((((......))))).))))). (-28.10 = -28.61 + 0.50)

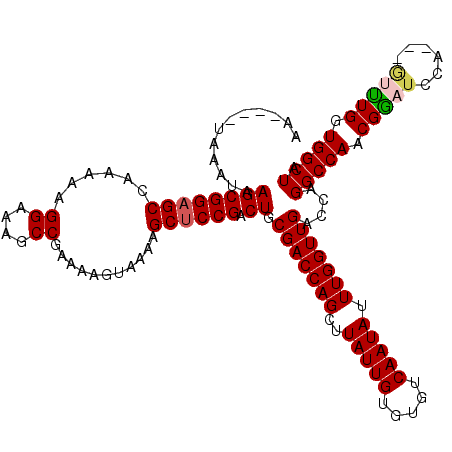
| Location | 17,300,525 – 17,300,638 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.81 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300525 113 + 23771897 UAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGAAUCCA---GUUUGGUGGCUAAGUGAGUGC----CUGGCCAGCGGGCGGAGGUGGAAUAAU ......(((((.((((..((((((((((.((.((((((.....))))))))(((((.(((((...---))))).))))).))))))...----))))...)))).))))).......... ( -38.80) >DroSim_CAF1 1038 113 + 1 UAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCA---GUUUGGUGGCUAAGUGAGUGG----CUGGCCAGCGGGCGGAGGUGGAAUAAU ......(((((.((((..((((((.(((.((.((((((.....))))))))(((((.(((((...---))))).))))).....)))))----))))...)))).))))).......... ( -40.00) >DroEre_CAF1 4723 112 + 1 UAAAAGCCCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCA----GUUGGUGGCUAAGUGAGUGC----CUGGCCAACGGGCGGAGGUGGAAUAAU ....((((((((((((((((((..(((((.....))))).))))))).((........))...))----))))).)))).......(((----(((.....))))))............. ( -37.70) >DroYak_CAF1 5056 120 + 1 UAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCACCAAACUGGUGGCUAAGUGAGUGCCUGCCUGGCCAGCGGGCGGAGGUGGAAUAAU ......(((((.((((((((((..(((((.....))))).))))))(.((((((..((..(((((((.....)))))....))..))....)))))).).)))).))))).......... ( -43.10) >consensus UAAAAGCUCCGACUGCGACCAGCUUAUUGUGUGUCAAUAUUUGGUUGACCAGGCCAACGGAUCCA___GUUUGGUGGCUAAGUGAGUGC____CUGGCCAGCGGGCGGAGGUGGAAUAAU ......(((((.((((..((((((((((.((.((((((.....))))))))(((((.(((((......))))).))))).)))))).......))))...)))).))))).......... (-34.06 = -34.81 + 0.75)

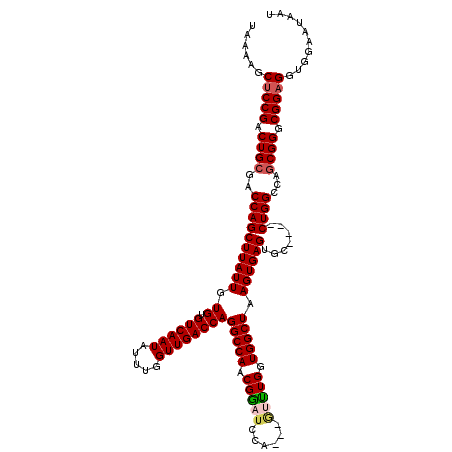
| Location | 17,300,525 – 17,300,638 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -27.75 |
| Energy contribution | -27.38 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300525 113 - 23771897 AUUAUUCCACCUCCGCCCGCUGGCCAG----GCACUCACUUAGCCACCAAAC---UGGAUUCGUUGGCCUGGUCAACCAAAUAUUGACACACAAUAAGCUGGUCGCAGUCGGAGCUUUUA ..........(((((.(.(((((((((----((....((....(((......---)))....))..)))))))))((((..(((((.....)))))...)))).)).).)))))...... ( -34.40) >DroSim_CAF1 1038 113 - 1 AUUAUUCCACCUCCGCCCGCUGGCCAG----CCACUCACUUAGCCACCAAAC---UGGAUCCGUUGGCCUGGUCAACCAAAUAUUGACACACAAUAAGCUGGUCGCAGUCGGAGCUUUUA ..........(((((.(.((.((((((----(.....(((..((((((....---.))......))))..)))........(((((.....))))).))))))))).).)))))...... ( -31.40) >DroEre_CAF1 4723 112 - 1 AUUAUUCCACCUCCGCCCGUUGGCCAG----GCACUCACUUAGCCACCAAC----UGGAUCCGUUGGCCUGGUCAACCAAAUAUUGACACACAAUAAGCUGGUCGCAGUCGGGGCUUUUA ..............(((((((((((((----((....((....(((.....----)))....))..)))))))))))....(((((.....))))).((((....))))..))))..... ( -35.00) >DroYak_CAF1 5056 120 - 1 AUUAUUCCACCUCCGCCCGCUGGCCAGGCAGGCACUCACUUAGCCACCAGUUUGGUGGAUCCGUUGGCCUGGUCAACCAAAUAUUGACACACAAUAAGCUGGUCGCAGUCGGAGCUUUUA ..........(((((.(.((.((((((.(((((....((....(((((.....)))))....))..)))))(((((.......)))))..........)))))))).).)))))...... ( -41.90) >consensus AUUAUUCCACCUCCGCCCGCUGGCCAG____GCACUCACUUAGCCACCAAAC___UGGAUCCGUUGGCCUGGUCAACCAAAUAUUGACACACAAUAAGCUGGUCGCAGUCGGAGCUUUUA ..........(((((.(.((.((((((..........(((..(((((((......)))......))))..)))........(((((.....)))))..)))))))).).)))))...... (-27.75 = -27.38 + -0.38)

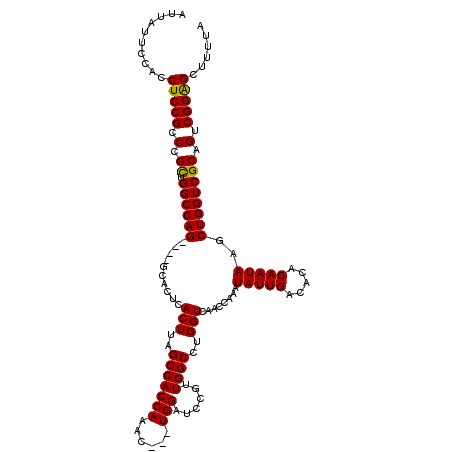

| Location | 17,300,565 – 17,300,678 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -32.65 |
| Energy contribution | -32.65 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300565 113 + 23771897 UUGGUUGACCAGGCCAACGAAUCCA---GUUUGGUGGCUAAGUGAGUGC----CUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCUGCCUGCAGUUUUACCGG .((((.((((((((.......(((.---(((((.((((((.((....))----.)))))).))))))))((((((((.((.(((....))).)).)))))))))))))..)))..)))). ( -38.30) >DroSim_CAF1 1078 113 + 1 UUGGUUGACCAGGCCAACGGAUCCA---GUUUGGUGGCUAAGUGAGUGG----CUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCUGCCUGCAGUUUUACCGG .((((.((((((((.......(((.---(((((.((((((.((.....)----))))))).))))))))((((((((.((.(((....))).)).)))))))))))))..)))..)))). ( -37.30) >DroEre_CAF1 4763 112 + 1 UUGGUUGACCAGGCCAACGGAUCCA----GUUGGUGGCUAAGUGAGUGC----CUGGCCAACGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCCGCCUGCAGUUUUACCGG .((((.((((((((.......(((.----(((.(((((((.((....))----.))))).)).))))))((((((((.((.(((....))).)).)))))))))))))..)))..)))). ( -37.70) >DroYak_CAF1 5096 120 + 1 UUGGUUGACCAGGCCAACGGAUCCACCAAACUGGUGGCUAAGUGAGUGCCUGCCUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCCGCCUGCAGUUUUACCGG .((((((..((((((.((..(.(((((.....))))).)..))..).)))))..))))))((((((...((((((((.((.(((....))).)).))))))))))))))........... ( -43.30) >consensus UUGGUUGACCAGGCCAACGGAUCCA___GUUUGGUGGCUAAGUGAGUGC____CUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCCGCCUGCAGUUUUACCGG .((((.(((..(((((......(((......)))..(((.....))).......))))).((((((...((((((((.((.(((....))).)).)))))))))))))).)))..)))). (-32.65 = -32.65 + -0.00)

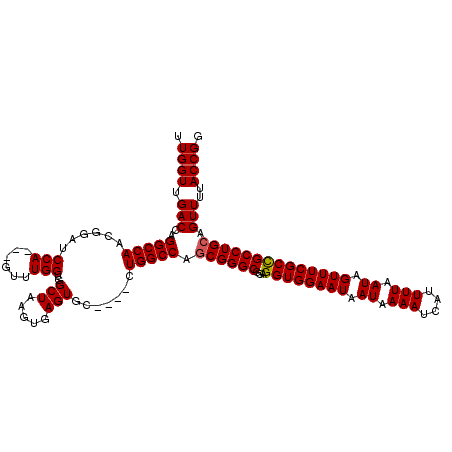
| Location | 17,300,602 – 17,300,718 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.90 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -28.96 |
| Energy contribution | -28.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17300602 116 + 23771897 AGUGAGUGC----CUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCUGCCUGCAGUUUUACCGGGCUUUAUGCAGCUCAAUGAAAUUAAAAUGAACUGUCAUUA .(((((.((----((((...((((((...((((((((.((.(((....))).)).)))))))))))))).......)))))))))))((((.(((.(........).))).))))..... ( -32.10) >DroSim_CAF1 1115 116 + 1 AGUGAGUGG----CUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCUGCCUGCAGUUUUACCGGGCUUUAUGCAGCUCAAUGAAAUUAAAAUGAACUGUCAUUA ....(((((----(.(..((((((((...((((((((.((.(((....))).)).))))))))))))))(((((((..(((((......)))))..)))))))....))..).)))))). ( -33.40) >DroEre_CAF1 4799 116 + 1 AGUGAGUGC----CUGGCCAACGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCCGCCUGCAGUUUUACCGGGCUUUACGGAGCUCAAUGAAAUUAAAAUGAACUGUCAUUA .(((((.((----((((..(((..((((.((((((((.((.(((....))).)).))))))))..)))).)))...)))))))))))..................(((((....))))). ( -33.80) >DroYak_CAF1 5136 120 + 1 AGUGAGUGCCUGCCUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCCGCCUGCAGUUUUACCGGGCUUUAUGGAGCUCAAUGAAAUUAAAAUGAACUGUCAUUA ..(((((.((.((((((..(((..((((.((((((((.((.(((....))).)).))))))))..)))).)))...)))))).....)).)))))..........(((((....))))). ( -37.70) >consensus AGUGAGUGC____CUGGCCAGCGGGCGGAGGUGGAAUAAUAAAAUCAUUUUAAUAGUUUCGCCGCCUGCAGUUUUACCGGGCUUUAUGCAGCUCAAUGAAAUUAAAAUGAACUGUCAUUA ..(((((........((((.((((((...((((((((.((.(((....))).)).)))))))))))))).(......).)))).......)))))..........(((((....))))). (-28.96 = -28.96 + 0.00)

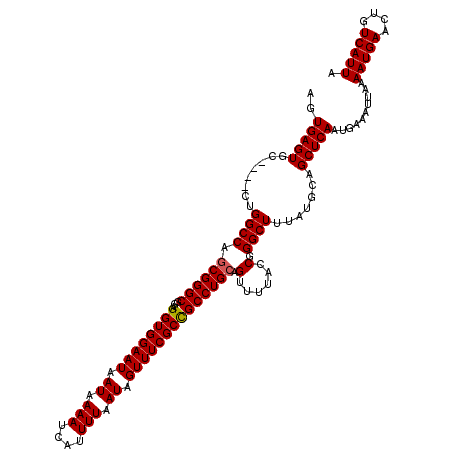
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:08 2006