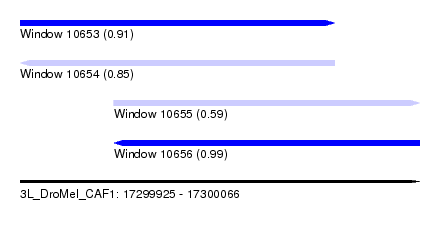
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,299,925 – 17,300,066 |
| Length | 141 |
| Max. P | 0.994145 |

| Location | 17,299,925 – 17,300,036 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -26.07 |
| Energy contribution | -27.82 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17299925 111 + 23771897 CCAGUUGAAGUUGGAAAACUGCUGCUCCUGCUG---------CCCGCCCAAUAGCAAAAUGGUAAAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUU (((((...((((....))))...))((((((((---------((.((......)).....)))....(((((....))))))))).)))........(((((((....)))))))))).. ( -31.30) >DroSim_CAF1 423 120 + 1 CCAGUUGAAGUUGGAAAACUGCUGCUCCUGCUGCUACUGCUGUCCGCCCAAUAGCAAAAUGGCAAAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUU (((((...((((....))))...))(((((((((((.((((((.......))))))...))))....(((((....))))))))).)))........(((((((....)))))))))).. ( -38.50) >DroEre_CAF1 4139 99 + 1 CCAGUUGAAGUUGGAAAACUGCUGCUCCUGCUG-------------UCCAAUAGCA--------AAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUU (((..(((((((....))))..(((((..((((-------------(....((((.--------.....)))).))))).))))).(....))))..(((((((....)))))))))).. ( -32.80) >DroYak_CAF1 4475 99 + 1 CCAGUUGAAGUUGGAAAACUGCUGCUCCUGCAG-------------CCCAAUAGCA--------AAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUU .((((((.((((....))))(((((....))))-------------)....((((.--------.....))))..))))))(((..(....).....(((((((....)))))))..))) ( -32.70) >consensus CCAGUUGAAGUUGGAAAACUGCUGCUCCUGCUG_____________CCCAAUAGCA________AAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUU (((..(((((((....))))..(((((.(((((..................)))))............((((....))))))))).(....))))..(((((((....)))))))))).. (-26.07 = -27.82 + 1.75)

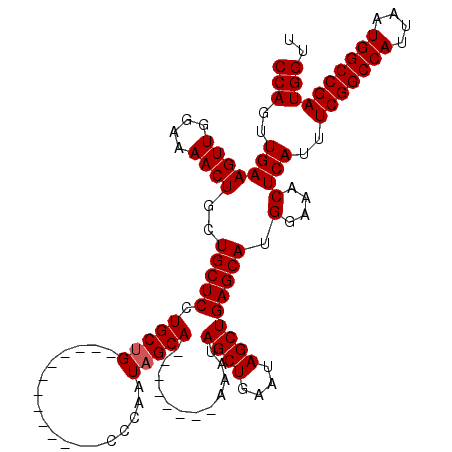
| Location | 17,299,925 – 17,300,036 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.852993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17299925 111 - 23771897 AACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUUUACCAUUUUGCUAUUGGGCGGG---------CAGCAGGAGCAGCAGUUUUCCAACUUCAACUGG .....(((((((....))))))).....(((((..(((((((((....)))).............(((....))))))---------))...)))))..(((((..........))))). ( -32.00) >DroSim_CAF1 423 120 - 1 AACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUUUGCCAUUUUGCUAUUGGGCGGACAGCAGUAGCAGCAGGAGCAGCAGUUUUCCAACUUCAACUGG .....(((((((....))))))).....(((.((.((((..((((((..(((..((((((............)))))).))))))))))))))))))..(((((..........))))). ( -36.00) >DroEre_CAF1 4139 99 - 1 AACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUU--------UGCUAUUGGA-------------CAGCAGGAGCAGCAGUUUUCCAACUUCAACUGG ..((.(((((((....)))))))....((((....(((((.(((.((((((.....--------.))...))))-------------.)))..)))))(.((((....)))).))))))) ( -28.70) >DroYak_CAF1 4475 99 - 1 AACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUU--------UGCUAUUGGG-------------CUGCAGGAGCAGCAGUUUUCCAACUUCAACUGG ..((((((((((....)))))))..(((((......))))).......(((.....--------.)))..)))(-------------((((....)))))......(((........))) ( -31.40) >consensus AACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUU________UGCUAUUGGG_____________CAGCAGGAGCAGCAGUUUUCCAACUUCAACUGG ..((.(((((((....)))))))....((((....(((((((((....))))............(((......................))).)))))(.((((....)))).))))))) (-25.52 = -25.53 + 0.00)

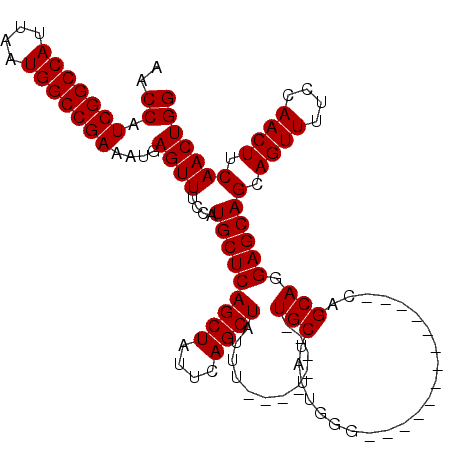
| Location | 17,299,958 – 17,300,066 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17299958 108 + 23771897 --CCCGCCCAAUAGCAAAAUGGUAAAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUUUGCGGAGUAACAAGGCGGAGAGUUGGC----------AGU --.(((((...................(((((....))))).((.((.(((((....(((((((....))))))).))))).))..)).....))))).........----------... ( -31.30) >DroSim_CAF1 463 110 + 1 UGUCCGCCCAAUAGCAAAAUGGCAAAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUUUGCGGAGUAACAAGGCGGAGAGUUGGC----------AGU (.((((((...((((.....(((......))).....)))).((.((.(((((....(((((((....))))))).))))).))..)).....)))))).)......----------... ( -34.70) >DroEre_CAF1 4172 106 + 1 ------UCCAAUAGCA--------AAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUUUGCGGAGUAACAAGGCGGAGAGUUGGUAGGGGUUGCUAGU ------(((....((.--------...(((((....))))).))..(.(((((....(((((((....))))))).))))).))))(((((...((.((...)).))....))))).... ( -29.70) >DroYak_CAF1 4508 96 + 1 ------CCCAAUAGCA--------AAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUUUGCGGAGUAACAAGGCGGAGAGUUGGU----------AGU ------.((((((((.--------.....))).....(((..((.((.(((((....(((((((....))))))).))))).))..)).....))).....))))).----------... ( -26.70) >consensus ______CCCAAUAGCA________AAAUAGCUGAAUAGCUGAGCAUGGAAACUCAUUUCGGCCAUUAAUGGCCGAUGGUUUGCGGAGUAACAAGGCGGAGAGUUGGC__________AGU .......(((((.((............(((((....))))).((.((.(((((....(((((((....))))))).))))).))..))......)).....))))).............. (-26.45 = -26.45 + -0.00)

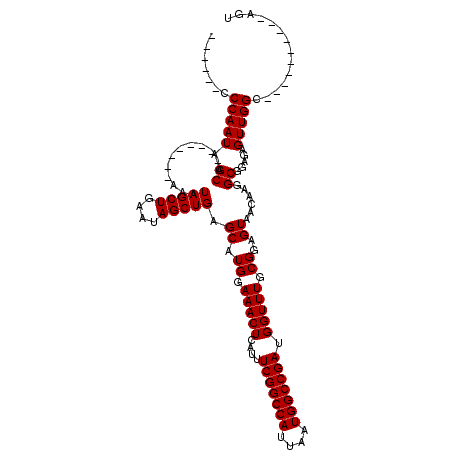
| Location | 17,299,958 – 17,300,066 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17299958 108 - 23771897 ACU----------GCCAACUCUCCGCCUUGUUACUCCGCAAACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUUUACCAUUUUGCUAUUGGGCGGG-- ...----------.........((((((.((......(((((...(((((((....)))))))...((((......))))((((....))))..........))))).)).)))))).-- ( -32.50) >DroSim_CAF1 463 110 - 1 ACU----------GCCAACUCUCCGCCUUGUUACUCCGCAAACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUUUGCCAUUUUGCUAUUGGGCGGACA ...----------........(((((((.((......(((((...(((((((....)))))))...((((......))))((((....))))..)))))......))....))))))).. ( -35.80) >DroEre_CAF1 4172 106 - 1 ACUAGCAACCCCUACCAACUCUCCGCCUUGUUACUCCGCAAACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUU--------UGCUAUUGGA------ ..............((((...................((......(((((((....)))))))..(((((......))))))).....(((.....--------.))).)))).------ ( -25.70) >DroYak_CAF1 4508 96 - 1 ACU----------ACCAACUCUCCGCCUUGUUACUCCGCAAACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUU--------UGCUAUUGGG------ ...----------.((((...................((......(((((((....)))))))..(((((......))))))).....(((.....--------.))).)))).------ ( -25.90) >consensus ACU__________ACCAACUCUCCGCCUUGUUACUCCGCAAACCAUCGGCCAUUAAUGGCCGAAAUGAGUUUCCAUGCUCAGCUAUUCAGCUAUUU________UGCUAUUGGG______ ..............((((...................((......(((((((....)))))))..(((((......))))))).....(((..............))).))))....... (-24.24 = -24.24 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:59 2006