| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,298,987 – 17,299,098 |
| Length | 111 |
| Max. P | 0.734558 |

| Location | 17,298,987 – 17,299,098 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -26.88 |
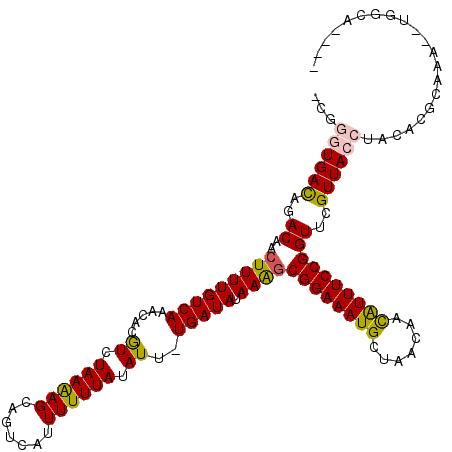
| Consensus MFE | -18.11 |
| Energy contribution | -17.62 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
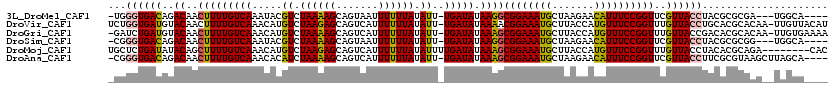
>3L_DroMel_CAF1 17298987 111 + 23771897 -UGGGUGACAGACAACUUUUGUCAAAUACGUCUAAAAGCAGUAAUUUUUUAUAUU-UGAUAUAAGGCGGAAAUGCUAAGAACAUUUCCGGUUCGUUACCUACGCGCGA---UGGCA---- -((((((((..((..(((.(((((((((....((((((.(....)))))))))))-))))).))).((((((((.......))))))))))..)))))))).((....---..)).---- ( -33.10) >DroVir_CAF1 79749 118 + 1 UCUGGUGAUGUACAACUUUUGUCAAACAUGUCUAAGAGCAGUCAUUUUUUAUAUU-UGAUAUAAAACGGAAAUGCUUACCAUGUUUCCGGUUUGUUACCUGCACGCACAA-UUGUUACAU .......(((((..((..((((((((((((.....(((((.((...((((((((.-..))))))))..))..)))))..)))))))..(((.....))).....).))))-..))))))) ( -20.60) >DroGri_CAF1 10255 117 + 1 -GAUCUGAUGUACAACUUUUGUCAAACAUGUCUAAAAGCAGUCAUUUUUUAUAUU-UGAUAUAAAGCGGAAAUGCUUACCAUGUUUCCGGUUUGUUACCGACACGCACAA-UUGUGAAAA -..................(((((((((((.....(((((.((...((((((((.-..))))))))..))..)))))..)))))))..(((.....)))))))((((...-.)))).... ( -20.80) >DroSim_CAF1 5985 111 + 1 -CGGGUGACAGACAACUUUUGUCAAAUACGUCUAAAAGCAGUAAUUUUUUAUAUU-UGAUAUAAGGCGGAAAUGCUAAGAACAUUUCCGGUUCGUUACCUACGCGCGG---UGGCA---- -.(((((((..((..(((.(((((((((....((((((.(....)))))))))))-))))).))).((((((((.......))))))))))..)))))))..((....---..)).---- ( -32.60) >DroMoj_CAF1 79756 112 + 1 UGCUCUGAUAUACAGCUUUUGUCAAACAUGUCUAAGAGCAGUCAUUUUUUAUAUUUUGAUAUAAAGCGGAAAUGCUUACCAUGUUUCCGGUUUGUUACCUACACGCAGA--------CAC (((((((((((...((....)).....)))))..))))))(((....(((((((....)))))))((((((((.........))))))(((.....))).....)).))--------).. ( -25.10) >DroAna_CAF1 13499 114 + 1 -CGGGUGACAGACAACUUUUGUCAAACACAUCUAAAAGCAGUCAUUUUUUAUAUU-UGAUAUAAAGCGGAAAUGCUAAGAACAUUUCCGGUUCGUUACCUUCGCGUAAGCUUAGCA---- -.(((((((..((..(((((((((((.......((((.......)))).....))-))))).))))((((((((.......))))))))))..)))))))..((....))......---- ( -29.10) >consensus _CGGGUGACAGACAACUUUUGUCAAACACGUCUAAAAGCAGUCAUUUUUUAUAUU_UGAUAUAAAGCGGAAAUGCUAACAACAUUUCCGGUUCGUUACCUACACGCAAA__UGGCA____ ...((((((..((..(((((((((.....((.((((((.......)))))).))..))))).))))((((((((.......))))))))))..))))))..................... (-18.11 = -17.62 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:55 2006