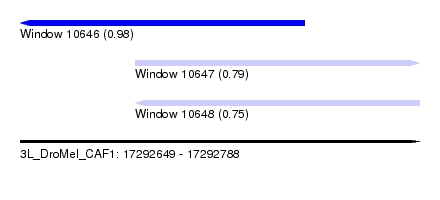
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,292,649 – 17,292,788 |
| Length | 139 |
| Max. P | 0.980008 |

| Location | 17,292,649 – 17,292,748 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.87 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17292649 99 - 23771897 CUGCAGCAGC---------------------AGCAACCACUGCAAAUGCAACUGCAACUGCAUGUGCCAGUCGGAGCAUCAUCAAUCCAGUGGUUCUCCACUCUCGACUCCUAAGCAGCG ((((.(((((---------------------((......))))..(((((........))))).))).(((((((..........)))(((((....)))))...)))).....)))).. ( -28.80) >DroSec_CAF1 176443 111 - 1 CUGCAGCAGCAG------UAGCAGCG---GCAGCAUCCACUGCAAAUGCAACUGCAACUGCAUCUGCCAGUCGGAGCAUCAUCAAUCCAGUGGUUCUCCACUCUCGACUCCUAAGCAGCG .(((((..((((------(.(((...---((((......))))...))).)))))..))))).((((.(((((((..........)))(((((....)))))...)))).....)))).. ( -41.10) >DroYak_CAF1 185213 120 - 1 CUGCAUCAUCAGUAGCUGCAACUGCAAGUGCAACUGCAAGUGCAAGUGGAACUGCAACUGCAACUGCCAGUCGGAGCAUCAUCAAUCCAGUGGUCCUCCACUCUCGACUCCUAAGUAGGA ((((.......((((.((((..((((..(.((..(((....)))..)).)..))))..)))).)))).(((((((..........)))(((((....)))))...)))).....)))).. ( -36.20) >consensus CUGCAGCAGCAG_______A_C_GC____GCAGCAACCACUGCAAAUGCAACUGCAACUGCAUCUGCCAGUCGGAGCAUCAUCAAUCCAGUGGUUCUCCACUCUCGACUCCUAAGCAGCG ((((.................................(((((((..(((....)))..)))).)))..(((((((..........)))(((((....)))))...)))).....)))).. (-22.31 = -22.87 + 0.56)


| Location | 17,292,689 – 17,292,788 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -26.40 |
| Energy contribution | -27.73 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17292689 99 + 23771897 GAUGCUCCGACUGGCACAUGCAGUUGCAGUUGCAUUUGCAGUGGUUGCU---------------------GCUGCUGCAGUGGGGGGUGCAUAAAUCACAGUCAUGCACGUGUUAAUAUC ((((((((.((((.((((.(((((.((..((((....))))..)).)))---------------------)))).)))))).))))((((((...........)))))).......)))) ( -35.60) >DroSec_CAF1 176483 111 + 1 GAUGCUCCGACUGGCAGAUGCAGUUGCAGUUGCAUUUGCAGUGGAUGCUGC---CGCUGCUA------CUGCUGCUGCAGUGGGGGGUGCAUAAAUCACAGUCAUGCACGUGUUAAUAUC ((((((((.((((.(((..(((((.(((((.(((...(((.....))))))---.))))).)------))))..))))))).))))((((((...........)))))).......)))) ( -45.20) >DroYak_CAF1 185253 120 + 1 GAUGCUCCGACUGGCAGUUGCAGUUGCAGUUCCACUUGCACUUGCAGUUGCACUUGCAGUUGCAGCUACUGAUGAUGCAGUGGGGGGUGCAUAAAUCACAGUCAUGCACGUGUUAAUAUC ((((((((.((((..(((((((..(((((...((.((((....)))).))...)))))..))))))).(....)...)))).))))((((((...........)))))).......)))) ( -38.80) >consensus GAUGCUCCGACUGGCAGAUGCAGUUGCAGUUGCAUUUGCAGUGGAUGCUGC____GC_G_U_______CUGCUGCUGCAGUGGGGGGUGCAUAAAUCACAGUCAUGCACGUGUUAAUAUC ((((((((.((((((((((((((......))))))))))(((....)))............................)))).))))((((((...........)))))).......)))) (-26.40 = -27.73 + 1.33)


| Location | 17,292,689 – 17,292,788 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17292689 99 - 23771897 GAUAUUAACACGUGCAUGACUGUGAUUUAUGCACCCCCCACUGCAGCAGC---------------------AGCAACCACUGCAAAUGCAACUGCAACUGCAUGUGCCAGUCGGAGCAUC ...........((((.((((((.(...((((((........((((...((---------------------((......))))...))))........))))))..)))))))..)))). ( -28.09) >DroSec_CAF1 176483 111 - 1 GAUAUUAACACGUGCAUGACUGUGAUUUAUGCACCCCCCACUGCAGCAGCAG------UAGCAGCG---GCAGCAUCCACUGCAAAUGCAACUGCAACUGCAUCUGCCAGUCGGAGCAUC ...........((((((((.......))))))))..((.(((((((..((((------(.((((..---((((((.....)))...)))..)))).)))))..))).)))).))...... ( -39.60) >DroYak_CAF1 185253 120 - 1 GAUAUUAACACGUGCAUGACUGUGAUUUAUGCACCCCCCACUGCAUCAUCAGUAGCUGCAACUGCAAGUGCAACUGCAAGUGCAAGUGGAACUGCAACUGCAACUGCCAGUCGGAGCAUC ...........((((.((((((((((..(((((........))))).))))((((.((((..((((..(.((..(((....)))..)).)..))))..)))).))))))))))..)))). ( -37.40) >consensus GAUAUUAACACGUGCAUGACUGUGAUUUAUGCACCCCCCACUGCAGCAGCAG_______A_C_GC____GCAGCAACCACUGCAAAUGCAACUGCAACUGCAUCUGCCAGUCGGAGCAUC ...........((((((((.......))))))))..((.((((((..................((....)).........((((..(((....)))..))))..)).)))).))...... (-19.83 = -20.33 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:51 2006