| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
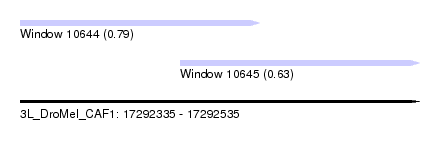
| Location | 17,292,335 – 17,292,535 |
| Length | 200 |
| Max. P | 0.792524 |

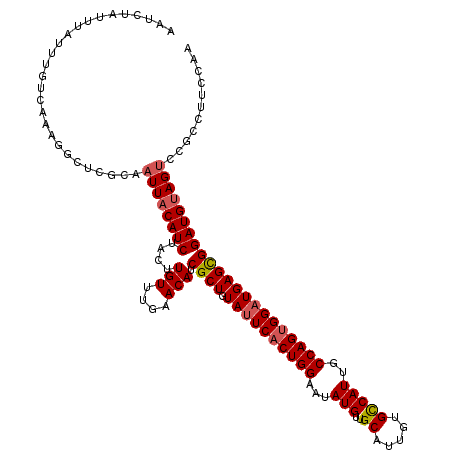
| Location | 17,292,335 – 17,292,455 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17292335 120 + 23771897 AAUCUAUUUAUUUGUCAAAGGCUCGCAAUUACAUCUACUUGUUUGAACAUCGCUGUAUUCCCUGGAAUAUGUUGCAUUGUGCCAUUGCCAGUGGAUGAGUGGAUGUAGUCCGCCUUCCAA .................(((((.....((((((((((((..(((.......((.((((((....))))))...))((((.((....)))))))))..))))))))))))..))))).... ( -33.60) >DroSec_CAF1 176138 110 + 1 AAUCUAUUUAUUUGUCAAAUGCUCGCAAUUACAUCUACUUGUUUGAACAUCGCUGUAUUCACUGGAAUAUGUUGCAUUGUGCCAUUGCCAGUGGAUGAGCGGAUGUAGUC---------- ...........................((((((((....(((....))).((((.((((((((((...(((..((.....)))))..)))))))))))))))))))))).---------- ( -29.20) >DroSim_CAF1 172356 120 + 1 AAUCUAUUUAUUUGUCAAAUGCUCGCAAUUACAUCUACUUGUUUGAACAUCGCUGUAUUCACUGGAAUAUGUUGCAUUGUGCCAUUGCCAGUGGAUGAGCGGAUGUAGUCCGCCUUCCAA ........................((.((((((((....(((....))).((((.((((((((((...(((..((.....)))))..))))))))))))))))))))))..))....... ( -31.30) >DroYak_CAF1 184905 120 + 1 AAUUCAUUUAUAAUUGAAAGGCUUGCAAUUACAUCUACUUGUUUGAACAUCGCUGUACUCACUGGAAUAUGCUGCAUUGUGUCAUUGCCAGUGGAUGAGUGGAUGCAGCCCGCCUUCCAA .............(((.(((((..((.....((((((((..((...(((....)))...(((((((((((((......)))).))).))))))))..))))))))..))..))))).))) ( -32.90) >consensus AAUCUAUUUAUUUGUCAAAGGCUCGCAAUUACAUCUACUUGUUUGAACAUCGCUGUAUUCACUGGAAUAUGUUGCAUUGUGCCAUUGCCAGUGGAUGAGCGGAUGUAGUCCGCCUUCCAA ...........................((((((((....(((....))).((((.((((((((((...(((..((.....)))))..))))))))))))))))))))))........... (-24.34 = -24.90 + 0.56)


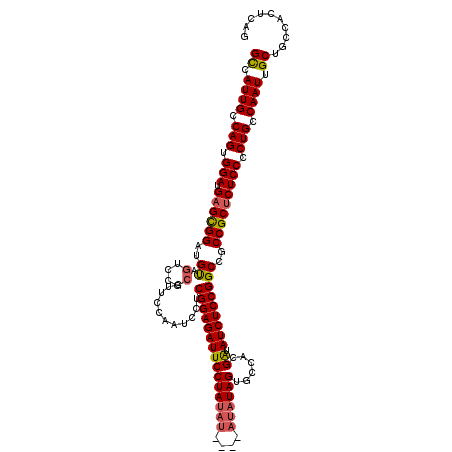
| Location | 17,292,415 – 17,292,535 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.99 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -30.54 |
| Energy contribution | -30.97 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17292415 120 + 23771897 GCCAUUGCCAGUGGAUGAGUGGAUGUAGUCCGCCUUCCAAUCCUCGGAGAUUCCUAUAUAUAUAUAUAGUACCACGAUAUCUCCGGCCGCCGCUCUCCCCUGCCAAUUGCUGCCACUCCG ((.((((.(((.(((.((((((..((.(....)...........(((((((((((((((....))))))......)).)))))))))..))))))))).))).)))).)).......... ( -33.70) >DroSec_CAF1 176218 105 + 1 GCCAUUGCCAGUGGAUGAGCGGAUGUAGUC-----------CCUCGGAGAUUCCUACAU----AUAUAGUGCCACGAUAUCUCCGGCCGCCGCUCUCCCCUGCCAAUUGCUGCCACUCAG ((.((((.(((.(((.((((((..((....-----------...(((((((((...(((----.....)))....)).)))))))))..))))))))).))).)))).)).......... ( -33.81) >DroSim_CAF1 172436 116 + 1 GCCAUUGCCAGUGGAUGAGCGGAUGUAGUCCGCCUUCCAAUCCUCGGAGAUUCCUAUAU----AUAUAGUGCCACGAUAUCUCCGGCCGCCGCUCUCCCCUGCCAAUUGCUGCCACUCAG ((.((((.(((.(((.((((((((((((......((((.......))))....))))))----.....(.(((...........)))).))))))))).))).)))).)).......... ( -33.40) >DroYak_CAF1 184985 114 + 1 GUCAUUGCCAGUGGAUGAGUGGAUGCAGCCCGCCUUCCAAUCCUCGGAGAUUCCUAU------AUAUAGUGCCACGGAAUCUCCGGCUGCCGCACUCCCCUGCCAAUUGCUGCCACUCAG ((.((((.(((.((..(((((...((((((.(.....).......(((((((((...------............)))))))))))))))..)))))))))).)))).)).......... ( -38.66) >consensus GCCAUUGCCAGUGGAUGAGCGGAUGUAGUCCGCCUUCCAAUCCUCGGAGAUUCCUAUAU____AUAUAGUGCCACGAUAUCUCCGGCCGCCGCUCUCCCCUGCCAAUUGCUGCCACUCAG ((.((((.(((.(((.((((((..((.(....)...........(((((((((((((((....))))))......)).)))))))))..))))))))).))).)))).)).......... (-30.54 = -30.97 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:49 2006