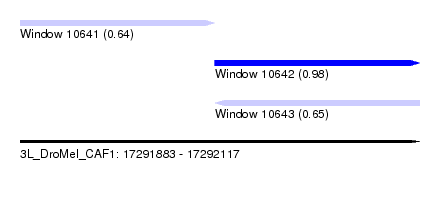
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,291,883 – 17,292,117 |
| Length | 234 |
| Max. P | 0.984925 |

| Location | 17,291,883 – 17,291,997 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17291883 114 + 23771897 CGACCCACCUUGCAACACCACCAUUUCCAUGCCACC------CCUUCCAUUGGCAGCACAAUGUGCGUAUAAAUAACGGAUUUCCUGUUAUAAAAUAUUUGUAACGACAUUUUCCCGAUU .......(.((((((.(((((.(((....(((((..------........)))))....)))))).))....(((((((.....))))))).......)))))).).............. ( -17.70) >DroSec_CAF1 175704 112 + 1 --ACCCACCUUGCAACACCACCAUCUCCAUGCCACC------CCAUCCACUGGCAGCACAAUGUGCGUAUAAAUAACGGAUUUCCUGUUAUAAAAUAUUUGUAACGACAUUUUCCCGAUU --.....(.((((((..............(((((..------........)))))((((...))))......(((((((.....))))))).......)))))).).............. ( -17.60) >DroSim_CAF1 171922 112 + 1 --ACCCACCUUGCAACGCCACCAUCUCCAUGCCACC------CCAUCCACUGGCAGCACAAUGUGCGUAUAAAUAACGGAUUUCCUGUUAUAAAAUAUUUGUAACGACAUUUUCCCGAUU --.....(.(((((((((...(((.....(((((..------........))))).....))).))))....(((((((.....)))))))........))))).).............. ( -19.10) >DroYak_CAF1 184437 118 + 1 --ACCCACCUUGCCACACCCCCAUUUCCAUGCCACCAUGCCACCAUCCACUGGCAGCACAAUGUGCGUAUAAAUAACGGAUUUCCUGUUAUAAAAUAUUUGUAACGACAUUUUCCCGAUU --..........................((((.....(((((........)))))((((...)))))))).......(((.....((((((((.....))))))))......)))..... ( -18.40) >consensus __ACCCACCUUGCAACACCACCAUCUCCAUGCCACC______CCAUCCACUGGCAGCACAAUGUGCGUAUAAAUAACGGAUUUCCUGUUAUAAAAUAUUUGUAACGACAUUUUCCCGAUU ..........(((................(((((................)))))((((...)))))))........(((.....((((((((.....))))))))......)))..... (-16.36 = -16.37 + 0.00)



| Location | 17,291,997 – 17,292,117 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -26.67 |
| Energy contribution | -26.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
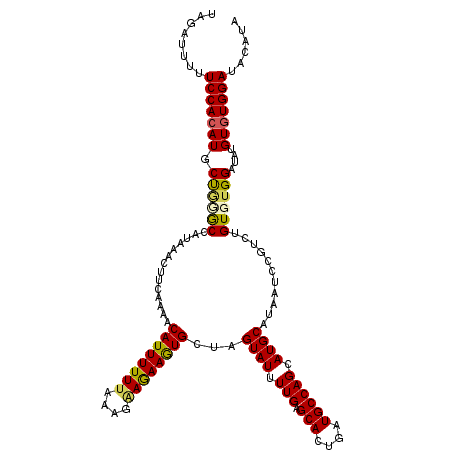
>3L_DroMel_CAF1 17291997 120 + 23771897 UAUGUAUCCACACAUAUCCACACAGACGGAUUAUGCAUUCUGGCAUCAGUGCUCAAGAUACUAGCAUUUCUUCUUUGAAAAAUGUUUUGAAGUUUGUAGCGUAGGAUGUGGAAAAAUCUA ......((((((....((((((((((((.(((((((......)))).))).)((((((((......((((......))))..)))))))).))))))...)).)))))))))........ ( -26.10) >DroSec_CAF1 175816 120 + 1 UAUGUAUCCACACAUAUCCACACAGACGGAUUAUGCAUGCUGGCAUCAGUGCUCAAAAUACUAGCACUUCUUCUUUAAAAAAUGUUUUGAAGUUUAUGGCCCAGCAUGUGGAAAAAUCGA ......(((((.(((((((........))).)))).((((((((...((((((.........))))))....((((((((....))))))))......).))))))))))))........ ( -32.10) >DroSim_CAF1 172034 120 + 1 UAUGUAUCCACACAUAUCCACACAGACGGAUUAUGCAUGCUGGCAUCAGUGCUCAAAAUACUAGCACUUCUUCUUUAAAAAAUGUUUUGAAGUUUAUGGCCCAGCAUGUGGAAAAAUCUA ......(((((.(((((((........))).)))).((((((((...((((((.........))))))....((((((((....))))))))......).))))))))))))........ ( -32.10) >DroYak_CAF1 184555 120 + 1 UAUGUAUCCACACAUAUCCACACAAACGGAUUAUGCAUGCUGGCAUCCGUGCUCAAAAUACUAGCACUUUUCCUUUAAAAAAUGUUUUCAAGUGUACUGUAAGGCAUUUGGAAAAACCUA .((((...((..(((((((........))).))))..))...))))..(((((.........))))).................(((((((((((........)))))))))))...... ( -22.40) >consensus UAUGUAUCCACACAUAUCCACACAGACGGAUUAUGCAUGCUGGCAUCAGUGCUCAAAAUACUAGCACUUCUUCUUUAAAAAAUGUUUUGAAGUUUAUGGCCCAGCAUGUGGAAAAAUCUA ......(((((.(((((((........))).)))).((((((((....(((((.........))))).....((((((((....))))))))......)).)))))))))))........ (-26.67 = -26.18 + -0.50)



| Location | 17,291,997 – 17,292,117 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17291997 120 - 23771897 UAGAUUUUUCCACAUCCUACGCUACAAACUUCAAAACAUUUUUCAAAGAAGAAAUGCUAGUAUCUUGAGCACUGAUGCCAGAAUGCAUAAUCCGUCUGUGUGGAUAUGUGUGGAUACAUA ........(((((((.((((((.((...........(((((((......)))))))...((((..((.(((....)))))..)))).......))..))))))....)))))))...... ( -26.30) >DroSec_CAF1 175816 120 - 1 UCGAUUUUUCCACAUGCUGGGCCAUAAACUUCAAAACAUUUUUUAAAGAAGAAGUGCUAGUAUUUUGAGCACUGAUGCCAGCAUGCAUAAUCCGUCUGUGUGGAUAUGUGUGGAUACAUA ........((((((((((((........((((...............)))).((((((.........))))))....)))))))(((((.((((......))))))))))))))...... ( -33.86) >DroSim_CAF1 172034 120 - 1 UAGAUUUUUCCACAUGCUGGGCCAUAAACUUCAAAACAUUUUUUAAAGAAGAAGUGCUAGUAUUUUGAGCACUGAUGCCAGCAUGCAUAAUCCGUCUGUGUGGAUAUGUGUGGAUACAUA ........((((((((((((........((((...............)))).((((((.........))))))....)))))))(((((.((((......))))))))))))))...... ( -33.86) >DroYak_CAF1 184555 120 - 1 UAGGUUUUUCCAAAUGCCUUACAGUACACUUGAAAACAUUUUUUAAAGGAAAAGUGCUAGUAUUUUGAGCACGGAUGCCAGCAUGCAUAAUCCGUUUGUGUGGAUAUGUGUGGAUACAUA ..(((...(((...(((.(((.(((((...(....)((((((((....))))))))...))))).)))))).))).)))..((((((((.((((......))))))))))))........ ( -27.10) >consensus UAGAUUUUUCCACAUGCUGGGCCAUAAACUUCAAAACAUUUUUUAAAGAAGAAGUGCUAGUAUUUUGAGCACUGAUGCCAGCAUGCAUAAUCCGUCUGUGUGGAUAUGUGUGGAUACAUA ........(((((((.((((((..............((((((((....))))))))...((((.(((.(((....)))))).))))...........))))))....)))))))...... (-22.36 = -22.55 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:47 2006