
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,285,718 – 17,285,878 |
| Length | 160 |
| Max. P | 0.965453 |

| Location | 17,285,718 – 17,285,838 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -48.42 |
| Consensus MFE | -43.01 |
| Energy contribution | -42.83 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17285718 120 + 23771897 UGGAUCUAUCUUGAUGGUCUCGGCCACGGCCUGAACGGCACUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGUAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUA ((((........((((((.(((..((..(((.....)))..))..)))..))))))(((((..(..(((((.....)))))..)..)))))..........)))).((((....)))).. ( -48.40) >DroSec_CAF1 169422 120 + 1 CGGAUCUAUCUUGAUGUUCUCGGCCACGGCCUGAACGGCGCUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGCAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUA .(((...........((((..(((....))).))))((((((((((....)..)))(((((..(.((((((.....)))))).)..)))))))))))....)))..((((....)))).. ( -49.20) >DroSim_CAF1 165641 120 + 1 CGGAUCUAUCUUGAUGGUCUCGGCCACGGCCUGAACGGCGCUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGCAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUA .(((........((((((.(((..((..(((.....)))..))..)))..))))))(((((..(.((((((.....)))))).)..)))))..........)))..((((....)))).. ( -49.00) >DroYak_CAF1 177656 120 + 1 GGGAUCUAUCUUGAUGGUCUCCGCCACGGCCUGAACAGCGCUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUGUCCAGGAGCGGCAGGAGUGCUCCCUUCCACUCCGCCCUCGGAUA ((((........(((((..(.((...((((((....)).))))..)).)..)))))((((((((..((((((...))))))..)))))))).....))))......((((....)))).. ( -47.10) >consensus CGGAUCUAUCUUGAUGGUCUCGGCCACGGCCUGAACGGCGCUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGCAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUA .(((.................(((....))).....((((((((((....)..)))((((((((.((((((.....)))))).))))))))))))))....)))..((((....)))).. (-43.01 = -42.83 + -0.19)



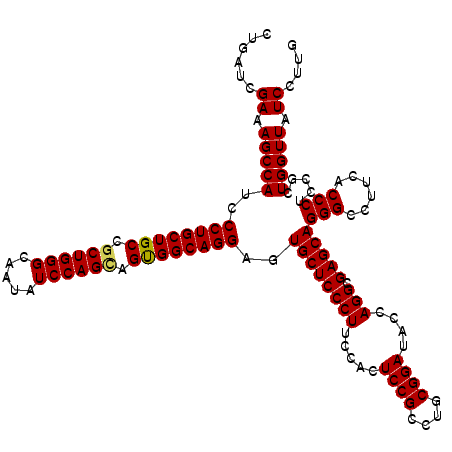
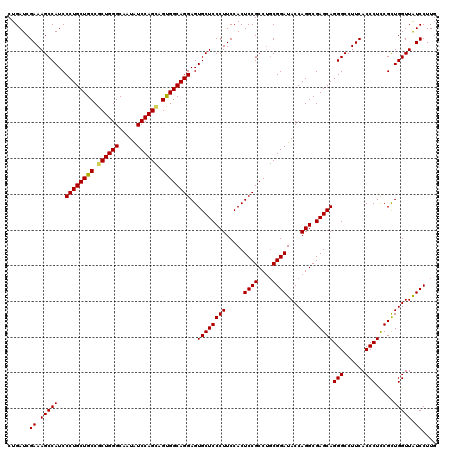
| Location | 17,285,758 – 17,285,878 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -48.64 |
| Consensus MFE | -47.49 |
| Energy contribution | -47.43 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17285758 120 + 23771897 CUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGUAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUACCAGGCGAGCAGGGCCUUCACCCUCCGCUGGUUAUCCUUG ......((.((((...(((((..(..(((((.....)))))..)..)))))((((((((((.....((((....))))....))).))))((((......))))..))))))).)).... ( -47.90) >DroSec_CAF1 169462 120 + 1 CUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGCAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUACCAGGCGAGCAGGGCCUUCACCCUCCGCUGGUUAUCCUUG ......((.((((...(((((..(.((((((.....)))))).)..)))))((((((((((.....((((....))))....))).))))((((......))))..))))))).)).... ( -49.00) >DroSim_CAF1 165681 120 + 1 CUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGCAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUACCAGGCGAGCAGGGCCUUCACCCUCCGCUGGUUAUCCUUG ......((.((((...(((((..(.((((((.....)))))).)..)))))((((((((((.....((((....))))....))).))))((((......))))..))))))).)).... ( -49.00) >DroYak_CAF1 177696 120 + 1 CUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUGUCCAGGAGCGGCAGGAGUGCUCCCUUCCACUCCGCCCUCGGAUACCAGGCGAGCAGGGCCUUCACCCUUCGUUGGUUGUCCUUC ..((..((.(((((.((((((((((.((((((...))))))(((.(((.((((((........))))))))))))........))).)))))))..............))))).))..)) ( -48.66) >consensus CUGAUCGAAAGCCAUCCCUGCUGCCGCUGGGCAAUAUCCAGCAGUGGCAGGAGUGCUCCCUUCCACUCCGCCUGCGGAUACCAGGCGAGCAGGGCCUUCACCCUCCGCUGGUUAUCCUUG ......((.(((((..((((((((.((((((.....)))))).))))))))..((((((((.....((((....))))....))).)))))(((......))).....))))).)).... (-47.49 = -47.43 + -0.06)

| Location | 17,285,758 – 17,285,878 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -51.27 |
| Consensus MFE | -49.17 |
| Energy contribution | -49.67 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
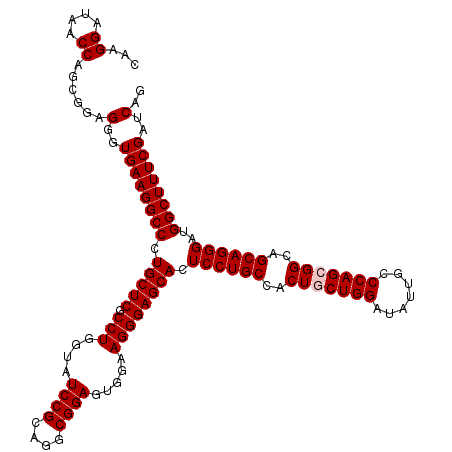
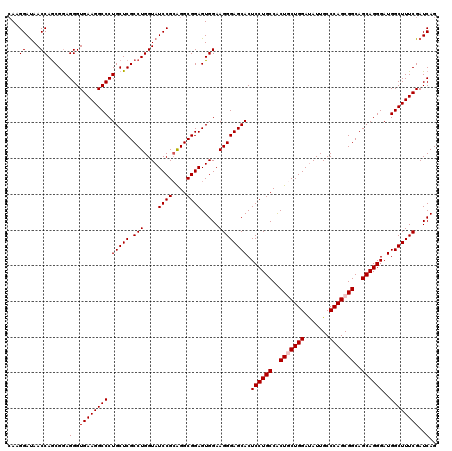
>3L_DroMel_CAF1 17285758 120 - 23771897 CAAGGAUAACCAGCGGAGGGUGAAGGCCCUGCUCGCCUGGUAUCCGCAGGCGGAGUGGAAGGGAGCACUCCUGCCACUACUGGAUAUUGCCCAGCGGCAGCAGGGAUGGCUUUCGAUCAG ....(((..(((..(.(((((....))))).)(((((((.......)))))))..)))..(..(((..((((((..(..((((.......))))..)..))))))...)))..).))).. ( -48.30) >DroSec_CAF1 169462 120 - 1 CAAGGAUAACCAGCGGAGGGUGAAGGCCCUGCUCGCCUGGUAUCCGCAGGCGGAGUGGAAGGGAGCACUCCUGCCACUGCUGGAUAUUGCCCAGCGGCAGCAGGGAUGGCUUUCGAUCAG ....(((..(((..(.(((((....))))).)(((((((.......)))))))..)))..(..(((..((((((..(((((((.......)))))))..))))))...)))..).))).. ( -53.60) >DroSim_CAF1 165681 120 - 1 CAAGGAUAACCAGCGGAGGGUGAAGGCCCUGCUCGCCUGGUAUCCGCAGGCGGAGUGGAAGGGAGCACUCCUGCCACUGCUGGAUAUUGCCCAGCGGCAGCAGGGAUGGCUUUCGAUCAG ....(((..(((..(.(((((....))))).)(((((((.......)))))))..)))..(..(((..((((((..(((((((.......)))))))..))))))...)))..).))).. ( -53.60) >DroYak_CAF1 177696 120 - 1 GAAGGACAACCAACGAAGGGUGAAGGCCCUGCUCGCCUGGUAUCCGAGGGCGGAGUGGAAGGGAGCACUCCUGCCGCUCCUGGACAUUGCCCAGCGGCAGCAGGGAUGGCUUUCGAUCAG ...((....))......(..((((((((.(((((.(((....((((....)))).....)))))))).((((((.(((.((((.......)))).))).))))))..))))))))..).. ( -49.60) >consensus CAAGGAUAACCAGCGGAGGGUGAAGGCCCUGCUCGCCUGGUAUCCGCAGGCGGAGUGGAAGGGAGCACUCCUGCCACUGCUGGAUAUUGCCCAGCGGCAGCAGGGAUGGCUUUCGAUCAG ...((....))......(..((((((((.(((((.(((....((((....)))).....)))))))).((((((..(((((((.......)))))))..))))))..))))))))..).. (-49.17 = -49.67 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:40 2006