
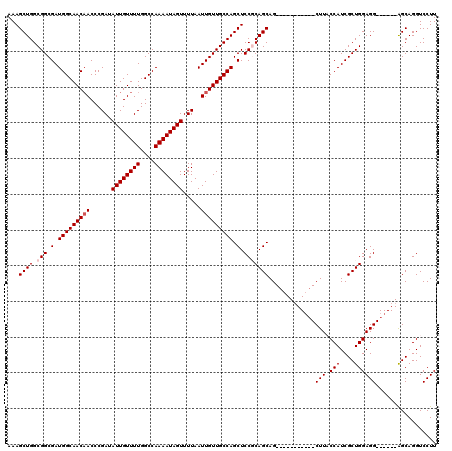
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,284,465 – 17,284,741 |
| Length | 276 |
| Max. P | 0.992039 |

| Location | 17,284,465 – 17,284,568 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.99 |
| Mean single sequence MFE | -37.74 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17284465 103 - 23771897 AAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG-----------CUUACCAUCGCUGGAGG------AGCAGGUCCUU .((((((((((.(.(((((((((......((((((((....)))))))).....))))))))).).))))..)))-----------))).(((....)))(((------(.....)))). ( -36.30) >DroSec_CAF1 168148 103 - 1 AAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG-----------CUUACCAUCGCUGGAGG------AGCAGGUCCUU .((((((((((.(.(((((((((......((((((((....)))))))).....))))))))).).))))..)))-----------))).(((....)))(((------(.....)))). ( -36.30) >DroSim_CAF1 164373 103 - 1 AAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG-----------CUUACCAUCGCUGGAGG------AGCAGGUCCUU .((((((((((.(.(((((((((......((((((((....)))))))).....))))))))).).))))..)))-----------))).(((....)))(((------(.....)))). ( -36.30) >DroEre_CAF1 171155 103 - 1 AAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCCCAGCAG-----------CUUUCCAUCGCUGGAGG------AGCAGGUCCUU ...(((((.((.(.(((((((((......((((((((....)))))))).....))))))))).).))))))).(-----------(((((((....)))).)------)))........ ( -38.40) >DroYak_CAF1 176321 120 - 1 AAAGCUGGCGGCGAUGGCAACCACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCCCAGCAGCUCCGCAGCUGCUUUCCAUCGCUGGAGGUCCUUUGGCAGGUCCUU ..((((((((((((((....).....((.((((((((....)))))))).)).)))))))))))))....(((((((....))))))).((((....))))((.(((.....))).)).. ( -41.40) >consensus AAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG___________CUUACCAUCGCUGGAGG______AGCAGGUCCUU ...((((.(((.(.(((((((((......((((((((....)))))))).....))))))))).).))))))).............(((.(((....))))))................. (-27.64 = -28.24 + 0.60)


| Location | 17,284,493 – 17,284,608 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -32.64 |
| Energy contribution | -33.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17284493 115 - 23771897 AACAACCCGCGCAACGAGCAGCAAACAACUGGCAACUGUGAAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG----- ..........((......((((.....((.(....).))....))))((((.(.(((((((((......((((((((....)))))))).....))))))))).).)))).))..----- ( -36.00) >DroSec_CAF1 168176 115 - 1 AACAACCCGCGCAACGAGCAGCAAACAACUGGCAACUGUGAAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG----- ..........((......((((.....((.(....).))....))))((((.(.(((((((((......((((((((....)))))))).....))))))))).).)))).))..----- ( -36.00) >DroSim_CAF1 164401 115 - 1 AACAACCCGCGCAACGAGCAGCAAACAACUGGCAACUGUGAAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG----- ..........((......((((.....((.(....).))....))))((((.(.(((((((((......((((((((....)))))))).....))))))))).).)))).))..----- ( -36.00) >DroEre_CAF1 171183 106 - 1 A---------GCAGCGAGCAGCGAACAACUGGCAACUGUGAAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCCCAGCAG----- .---------((((....(((.......)))....))))....(((((.((.(.(((((((((......((((((((....)))))))).....))))))))).).)))))))..----- ( -34.70) >DroYak_CAF1 176361 120 - 1 AAGAGCGCGCGCAACGAGCAGCAAACAACUGGCAACUGUGAAAGCUGGCGGCGAUGGCAACCACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCCCAGCAGCUCCG ..((((((..((.....)).))...((...(....)..))...(((((.((.(.(((((((........((((((((....)))))))).......))))))).).))))))).)))).. ( -39.46) >consensus AACAACCCGCGCAACGAGCAGCAAACAACUGGCAACUGUGAAAGCUGGCGGCGAUGGCAACAACCCGAUAUUGUUUUGGCCAAAAUAGUUUUAAUUGUUGCCAGCUCCGCAGCAG_____ ..........((......((((.....((.(....).))....))))((((.(.(((((((((......((((((((....)))))))).....))))))))).).)))).))....... (-32.64 = -33.24 + 0.60)



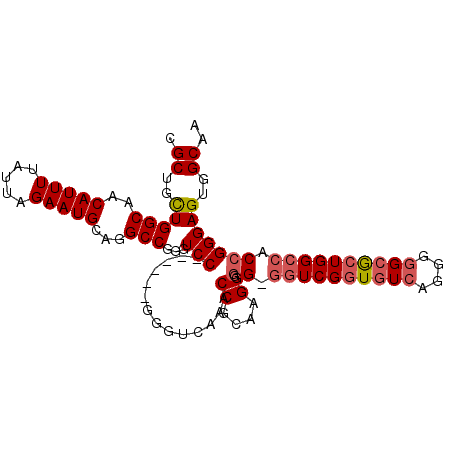
| Location | 17,284,608 – 17,284,703 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -32.53 |
| Energy contribution | -33.65 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17284608 95 + 23771897 CGCUGUUGGCAACAUUUUAUUAGAAUGCAGGCCGGGUCC------GGGUCAAACCGCAAGGG-G-GGUCGGUGUCAAGGAGCGCUGGCCACCGGGAGUGGCAA .((..(((((..(((((.....)))))...)))...(((------((......((....)).-.-(((((((((......))))))))).)))))))..)).. ( -39.30) >DroSec_CAF1 168291 96 + 1 CGCUGCUGGCAACAUUUUAUUAGAAUGCAGGCCGGGUCC------GGGUCAAACCGCAAGGGGG-GGUCGGUGUCAGGGGGCGCUGGCCACCGGGAGUGGCAA .((..(((((..(((((.....)))))...)))...(((------((......((....))...-((((((((((....)))))))))).)))))))..)).. ( -44.60) >DroSim_CAF1 164516 96 + 1 CGCUGCUGGCAACAUUUUAUUAGAAUGCAGGCCGGGUCC------GGGUCAAACCGCAAGGGGG-GGUCGGUGUCAGGGGGCGCUGGCCACCGGGAGUGGCAA .((..(((((..(((((.....)))))...)))...(((------((......((....))...-((((((((((....)))))))))).)))))))..)).. ( -44.60) >DroYak_CAF1 176481 103 + 1 CGCUGCUGGCAACAUUUUAUUAGAAUGCAGGCCGAGUCCGGGUACGGGUCAAACCGCAAGGGGGGAUUCGGUGUCAGGGGGGAGUGGCCACCGGGAGUGGCAA ((((.((((...(((((.....)))))..(((((..(((.(..(((((((...((....))...))))).))..)....)))..))))).)))).)))).... ( -40.20) >consensus CGCUGCUGGCAACAUUUUAUUAGAAUGCAGGCCGGGUCC______GGGUCAAACCGCAAGGGGG_GGUCGGUGUCAGGGGGCGCUGGCCACCGGGAGUGGCAA .((..(((((..(((((.....)))))...)))...(((..............((....)).((.((((((((((....)))))))))).)))))))..)).. (-32.53 = -33.65 + 1.12)

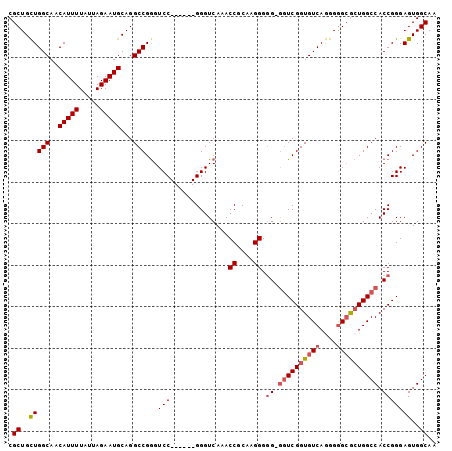
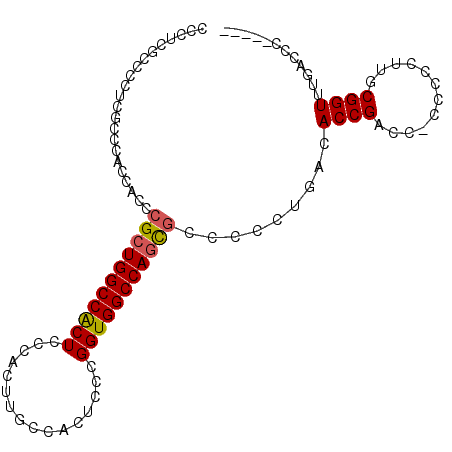
| Location | 17,284,647 – 17,284,741 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17284647 94 - 23771897 CCCUAACCCCUCGCCCAACACCCGCUGGCCACUCCCACUUGCCACUCCCGGUGGCCAGCGCUCCUUGACACCGACC-C-CCCUUGCGGUUUGACCC----- ...............(((....(((((((((((................)))))))))))....)))..((((...-.-......)))).......----- ( -23.49) >DroSec_CAF1 168330 95 - 1 CCUUCGCCCCUCGCCCACCACCCGCUGGCCGCUCCCACUUGCCACUCCCGGUGGCCAGCGCCCCCUGACACCGACC-CCCCCUUGCGGUUUGACCC----- ......................(((((((((((................))))))))))).........((((.(.-.......))))).......----- ( -22.39) >DroSim_CAF1 164555 95 - 1 CCUUCGCCCCUCGCCCACCACCCGCUGGCCGCUCCCACUUGCCACUCCCGGUGGCCAGCGCCCCCUGACACCGACC-CCCCCUUGCGGUUUGACCC----- ......................(((((((((((................))))))))))).........((((.(.-.......))))).......----- ( -22.39) >DroYak_CAF1 176521 83 - 1 --CU----------------CCUGCUGGCCACUCCCACUUGCCACUCCCGGUGGCCACUCCCCCCUGACACCGAAUCCCCCCUUGCGGUUUGACCCGUACC --..----------------.....((((((((................))))))))..........................(((((......))))).. ( -16.69) >consensus CCCUCGCCCCUCGCCCACCACCCGCUGGCCACUCCCACUUGCCACUCCCGGUGGCCAGCGCCCCCUGACACCGACC_CCCCCUUGCGGUUUGACCC_____ ......................(((((((((((................))))))))))).........((((............))))............ (-20.05 = -20.11 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:37 2006