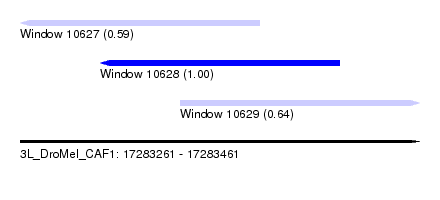
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,283,261 – 17,283,461 |
| Length | 200 |
| Max. P | 0.997771 |

| Location | 17,283,261 – 17,283,381 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -55.00 |
| Consensus MFE | -49.18 |
| Energy contribution | -50.98 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17283261 120 - 23771897 CCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGAACUUUGUGCGGCCACGUUGCACGUCCACGACCCCCCCUGC ..((((..((.((((..((((((....))))))((((.((..(((......)))..)).))))(((((.(((((((.........))))))))))))....)).)).))..))))..... ( -55.20) >DroSec_CAF1 166970 114 - 1 CCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGAACUUUGUGCGGCCACGUUGCACGUCCACGUCCCC------ ..((((.(((.((((..((((((....))))))((((.((..(((......)))..)).))))(((((.(((((((.........))))))))))))....)).)).)))))))------ ( -57.00) >DroSim_CAF1 163178 114 - 1 CCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGAACUUUGUGCGGCCACGUUGCACGUCCACGUCCCC------ ..((((.(((.((((..((((((....))))))((((.((..(((......)))..)).))))(((((.(((((((.........))))))))))))....)).)).)))))))------ ( -57.00) >DroEre_CAF1 170036 111 - 1 CCGGGGCACGAGGCGAGUCCUGUAGCAACAGG---------AGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGAACUUUGUGCGGCCACGUUGCACGUCCACAUCCCCCCAUGC ..((((...((..((..((((((....)))))---------).))...))..((((.((..(((((((.(((((((.........))))))))))).))).)).))))....)))).... ( -47.30) >DroYak_CAF1 175123 118 - 1 CCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGAACUUUGUGCGGCCACGUUGCACGUCCACGUCCCCCU--UC ..((((.(((.((((..((((((....))))))((((.((..(((......)))..)).))))(((((.(((((((.........))))))))))))....)).)).))).)))).--.. ( -58.50) >consensus CCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGAACUUUGUGCGGCCACGUUGCACGUCCACGUCCCCC____C ..((((.(((.((((..((((((....))))))((((.((..(((......)))..)).))))(((((.(((((((.........))))))))))))....)).)).)))))))...... (-49.18 = -50.98 + 1.80)

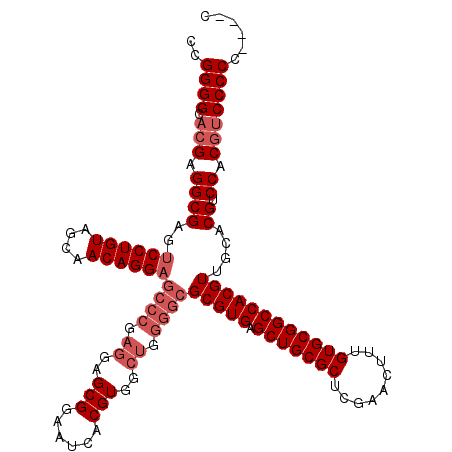
| Location | 17,283,301 – 17,283,421 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -50.04 |
| Consensus MFE | -45.00 |
| Energy contribution | -46.60 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17283301 120 - 23771897 GCAAAAACGGCACCUUUGGGAAAUUAAUAUCCUUUUUGGGCCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGA ((.....((((.((...((((........))))....))))))..)).(((((((..((((((....))))))((((.((..(((......)))..)).))))((....))))).)))). ( -52.40) >DroSec_CAF1 167004 120 - 1 GCAAAAACGGCACCUUUGGGAAAUUAAUAUGCUUUCUGGGCCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGA ((.....((((.((...(..(.((....))..)..).))))))..)).(((((((..((((((....))))))((((.((..(((......)))..)).))))((....))))).)))). ( -48.30) >DroSim_CAF1 163212 120 - 1 GCAAAAACGGCACCUUUGGGAAAUUAAUAUCCUUUUUGGGCCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGA ((.....((((.((...((((........))))....))))))..)).(((((((..((((((....))))))((((.((..(((......)))..)).))))((....))))).)))). ( -52.40) >DroEre_CAF1 170076 111 - 1 GCAAAAACGGCACCUUUGGGAAAUUAAUAUCCUUUUUGGGCCGGGGCACGAGGCGAGUCCUGUAGCAACAGG---------AGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGA ((.....((((.((...((((........))))....))))))..)).((((.(...((((((....)))))---------)((((..(((((((.(...).))))))).))))))))). ( -44.70) >DroYak_CAF1 175161 120 - 1 GCAAAAACGGCACCUUUGGGAAAUUAAUAUCCUUUUUGGGCCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGA ((.....((((.((...((((........))))....))))))..)).(((((((..((((((....))))))((((.((..(((......)))..)).))))((....))))).)))). ( -52.40) >consensus GCAAAAACGGCACCUUUGGGAAAUUAAUAUCCUUUUUGGGCCGGGGCACGAGGCGAGUCCUGUAGCAACAGGAGCCCGAGGAGCGGAAUCACGUGGCUGGGGCGCGUGAGCUGCGCUCGA ((.....((((.((...((((........))))....))))))..)).(((((((..((((((....))))))((((.((..(((......)))..)).))))((....))))).)))). (-45.00 = -46.60 + 1.60)

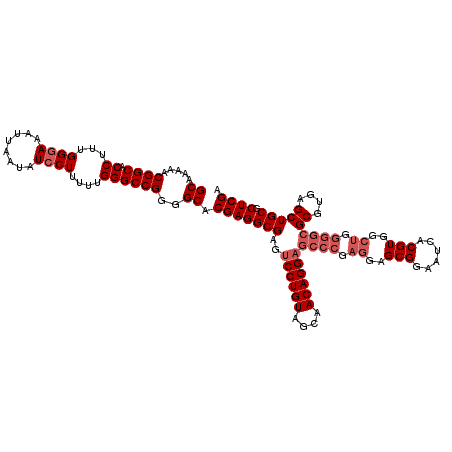
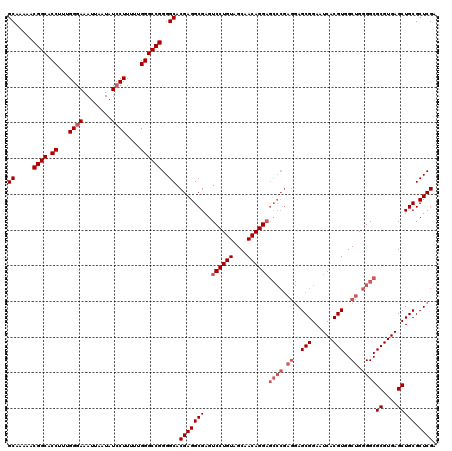
| Location | 17,283,341 – 17,283,461 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -31.64 |
| Energy contribution | -33.64 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17283341 120 + 23771897 CUCGGGCUCCUGUUGCUACAGGACUCGCCUCGUGCCCCGGCCCAAAAAGGAUAUUAAUUUCCCAAAGGUGCCGUUUUUGCUGGCGAAAAUCACGCAAAAUGCGACCCGGCUUUCACGAGU ..((((.((((((....)))))))))).((((((...((((((.....(((........)))....)).)))).....(((((.(.....).(((.....)))..)))))...)))))). ( -40.90) >DroSec_CAF1 167044 120 + 1 CUCGGGCUCCUGUUGCUACAGGACUCGCCUCGUGCCCCGGCCCAGAAAGCAUAUUAAUUUCCCAAAGGUGCCGUUUUUGCUGGCGAAAAUCACGCAAAAUUCGACCCGGCUUUCACGAGU ..((((.((((((....)))))))))).((((((...((((((.((((.........)))).....)).)))).....(((((((((............))))..)))))...)))))). ( -35.80) >DroSim_CAF1 163252 120 + 1 CUCGGGCUCCUGUUGCUACAGGACUCGCCUCGUGCCCCGGCCCAAAAAGGAUAUUAAUUUCCCAAAGGUGCCGUUUUUGCUGGCGAAAAUCACGCAAAAUGCGACCCGGCUUUCACGAGU ..((((.((((((....)))))))))).((((((...((((((.....(((........)))....)).)))).....(((((.(.....).(((.....)))..)))))...)))))). ( -40.90) >DroEre_CAF1 170115 112 + 1 --------CCUGUUGCUACAGGACUCGCCUCGUGCCCCGGCCCAAAAAGGAUAUUAAUUUCCCAAAGGUGCCGUUUUUGCUGGCGAAAAUCACGCAAAAUGCGACCCGGCUUUCAAGAGU --------(((((....)))))(((((((....((..((((((.....(((........)))....)).)))).....)).)))((((..(.(((.....)))....)..))))..)))) ( -32.30) >DroYak_CAF1 175201 120 + 1 CUCGGGCUCCUGUUGCUACAGGACUCGCCUCGUGCCCCGGCCCAAAAAGGAUAUUAAUUUCCCAAAGGUGCCGUUUUUGCUGGCAAAAAUCACGCAAAAUGCGACCCGGCUUUCAAGAGU ((((((.((((((....)))))))))(((((((....((((((.....(((........)))....)).)))).(((((((((......))).)))))).))))...)))......))). ( -37.50) >consensus CUCGGGCUCCUGUUGCUACAGGACUCGCCUCGUGCCCCGGCCCAAAAAGGAUAUUAAUUUCCCAAAGGUGCCGUUUUUGCUGGCGAAAAUCACGCAAAAUGCGACCCGGCUUUCACGAGU ..((((.((((((....)))))))))).((((((...((((((.....(((........)))....)).)))).....(((((.(.....).(((.....)))..)))))...)))))). (-31.64 = -33.64 + 2.00)

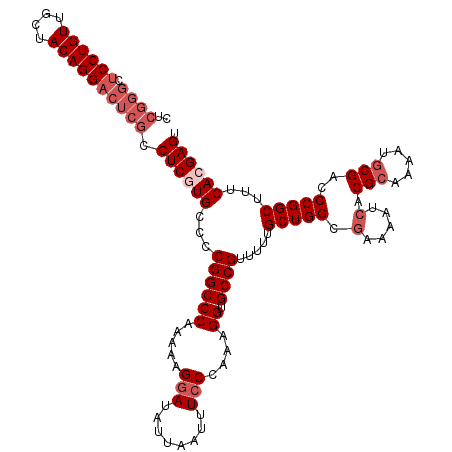
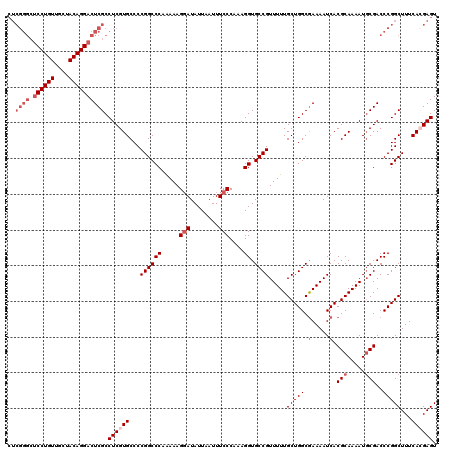
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:34 2006