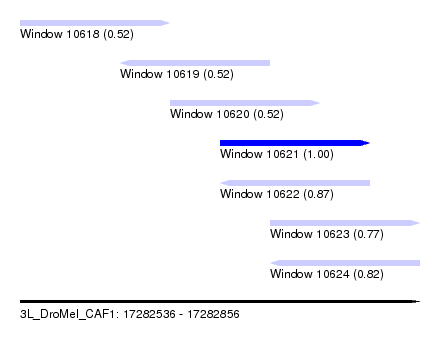
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,282,536 – 17,282,856 |
| Length | 320 |
| Max. P | 0.998784 |

| Location | 17,282,536 – 17,282,656 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.90 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -27.86 |
| Energy contribution | -29.28 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282536 120 + 23771897 GAUCUCUAUGGAAACGUGAAGCUGAGGCCGUAGCUGGAGCUGAAGCUGAAGCGCAGCUAAUAAGGUCUGCGGGCCUGCAGGGCGUAUGCUUAAUAUUUUGAGCCAGUUGGCGCAUAUAUA ...((.((((....)))).))((((((((...(((..(((....)))..)))((((((.....)).)))).))))).)))...((((((((((....))))(((....)))))))))... ( -42.70) >DroSec_CAF1 166263 118 + 1 G--CUCUAUGGAAACUUGAAGCUGAGGCCGUAGCUGGAGCUGACGCUGAGGGUCAGCUAAUAAGGUCUGCGGGCCUGCAGGGCGUAUUCUUAAUAUUUUGAGCCAGUUGGCGCUUAUAUA (--((.((.(....).)).)))((((((((..((((((((((((.(...).)))))))((((.(.((((((....)))))).).))))(((((....)))))))))))))).)))).... ( -40.60) >DroSim_CAF1 162449 118 + 1 G--CUCUAUGGAAACUUGAAGCUGAGGCCGUAGCUGGAGCUGAAGCUGAAGCGCAGCUAAUAAGGUCUGCGGGCCUGCAGGGCGUAUGCUUAAUAUUUUGAGCCAGUUGGCGCUUAUAUA (--((.((.(....).)).)))(((((((...(((..(((....)))..)))((((((.....)).)))).))))).))((((((..((((((....))))))......))))))..... ( -40.80) >DroEre_CAF1 169305 118 + 1 G--CUCUAUGGAAACUUGAAGCUGAGGCCAUAGCUGGAGCCGAAGCUGAAGCGCAGCUAAUAAGGUCUGCGGACCUGCGGGGCGUAUGCUUAAUAUUUUGAGCCAGUUGGCGCUUAUAUA (--((.((.(....).)).)))((((((((..(((((..(.(((....(((((..(((....(((((....)))))....)))...)))))....))).)..))))))))).)))).... ( -40.80) >DroYak_CAF1 174370 106 + 1 G--CUCUAUGGAAACUUGAAGCUAAGGCUGUAG------------CUAAAACGCAGCUAAUAAGGUCUGCGGAGCUGCAGGGCGUAUGCUUAAUAUUUUGAGCCAGUUGGCGCUUAUAUA (--((.((.(....).)).))).....((((((------------((....(((((((.....)).))))).))))))))(((((..((((((....))))))......)))))...... ( -37.00) >consensus G__CUCUAUGGAAACUUGAAGCUGAGGCCGUAGCUGGAGCUGAAGCUGAAGCGCAGCUAAUAAGGUCUGCGGGCCUGCAGGGCGUAUGCUUAAUAUUUUGAGCCAGUUGGCGCUUAUAUA ....((...(....)..))..((((((((.(((((........)))))...(((((((.....)).)))))))))).)))(((((..((((((....))))))......)))))...... (-27.86 = -29.28 + 1.42)



| Location | 17,282,616 – 17,282,736 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282616 120 - 23771897 AAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUUAUGCAGAACUUUCCGCUUUGGUGCUCUGGGCAACGGCAUUUAUAUAUGCGCCAACUGGCUCAAAAUAUUAAGCAUACGCC ...(((((((.....((((((..(((((....)))))..)))))))))))))..((....(((((...(....).((((......))))(((....)))...........)))))..)). ( -25.70) >DroSec_CAF1 166341 120 - 1 AAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUUAUGCAGAACUUUCCGCUUUGGUGCUCUGGGCAACGGCAUUUAUAUAAGCGCCAACUGGCUCAAAAUAUUAAGAAUACGCC ......((.(((.((((((((..(((((....)))))..)))))))).......((((((((((((((.....)))..........))))))))..)))............))).))... ( -27.10) >DroSim_CAF1 162527 120 - 1 AAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUUAUGCAGAACUUUACGCUUUGGUGCUCUGGGCAACGGCAUUUAUAUAAGCGCCAACUGGCUCAAAAUAUUAAGCAUACGCC ...(((((((.....((((((..(((((....)))))..)))))))))))))..((((.((((((...(....))))))).....))))(((....)))............((....)). ( -25.60) >DroEre_CAF1 169383 120 - 1 AAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUUAUGCAGAACUUUCCGCUUUGGUGCUCUGGCCAACGGCAUUUAUAUAAGCGCCAACUGGCUCAAAAUAUUAAGCAUACGCC ......((((...((((((((..(((((....)))))..)))))))).......(((((((((((...(((...))).........))))))))..)))..........))))....... ( -25.80) >DroYak_CAF1 174436 120 - 1 AAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUUAUGCAGAACUUUCCGCUUUGGCGCUCCGGCCAGCGGCAUUUAUAUAAGCGCCAACUGGCUCAAAAUAUUAAGCAUACGCC ......((((...((((((((..(((((....)))))..)))))))).......((((((((((((((.....)))..........))))))))..)))..........))))....... ( -28.10) >consensus AAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUUAUGCAGAACUUUCCGCUUUGGUGCUCUGGGCAACGGCAUUUAUAUAAGCGCCAACUGGCUCAAAAUAUUAAGCAUACGCC ...(((((((.....((((((..(((((....)))))..)))))))))))))..((((((((((((((.....)))..........))))))))..)))..................... (-25.04 = -24.92 + -0.12)



| Location | 17,282,656 – 17,282,776 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282656 120 + 23771897 AAUGCCGUUGCCCAGAGCACCAAAGCGGAAAGUUCUGCAUAAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGGGAAAUGCU ..(((..((((((...((......(((((....))))).............................(((((.....)))))..(((......))))).))))))..))).......... ( -30.00) >DroSec_CAF1 166381 120 + 1 AAUGCCGUUGCCCAGAGCACCAAAGCGGAAAGUUCUGCAUAAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCU ..(((..((((((...((......(((((....))))).............................(((((.....)))))..(((......))))).))))))..))).......... ( -30.00) >DroSim_CAF1 162567 120 + 1 AAUGCCGUUGCCCAGAGCACCAAAGCGUAAAGUUCUGCAUAAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCU ..(((..((((((...((.......((.((((((((((((....................)))))...))..)))))...))..(((......))))).))))))..))).......... ( -28.05) >DroEre_CAF1 169423 120 + 1 AAUGCCGUUGGCCAGAGCACCAAAGCGGAAAGUUCUGCAUAAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCU ...(((...)))...((((((...(((((....)))))........................((...(((((.....)))))..(((......))))))).((....)).......)))) ( -26.70) >DroYak_CAF1 174476 120 + 1 AAUGCCGCUGGCCGGAGCGCCAAAGCGGAAAGUUCUGCAUAAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCU ....((((.(((.....((((...(..(((((((((((((....................)))))...))..))))))..)...)))).....))))))).((....))........... ( -32.45) >consensus AAUGCCGUUGCCCAGAGCACCAAAGCGGAAAGUUCUGCAUAAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCU ..(((((((......))).((...(((((....)))))........................((...(((((.....)))))..(((......)))))))))))..((((......)))) (-25.78 = -25.82 + 0.04)

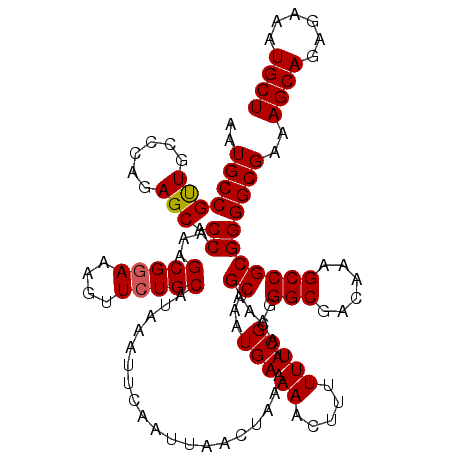

| Location | 17,282,696 – 17,282,816 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282696 120 + 23771897 AAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGGGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGA ...(((((((..(((......((((..((((.......((((..(((......))).))))((....)).(((...(((......))).))).))))..))))......))).))))))) ( -28.20) >DroSec_CAF1 166421 120 + 1 AAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGA ...(((((((..(((......((((..((((.......((((..(((......))).))))((....)).(((...(((......))).))).))))..))))......))).))))))) ( -29.10) >DroSim_CAF1 162607 120 + 1 AAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGA ...(((((((..(((......((((..((((.......((((..(((......))).))))((....)).(((...(((......))).))).))))..))))......))).))))))) ( -29.10) >DroEre_CAF1 169463 120 + 1 AAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGA ...(((((((..(((......((((..((((.......((((..(((......))).))))((....)).(((...(((......))).))).))))..))))......))).))))))) ( -29.10) >DroYak_CAF1 174516 120 + 1 AAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCACUCAGAUGCACAGAAGUGGCAAUUGGA ...(((((((..(((......((((..(((........((((..(((......))).))))((....)).(((...(((......))).)))..)))..))))......))).))))))) ( -28.00) >consensus AAAUUCAAUUAACUAAAAACAUGCAAAUGAAAACUUUUUUCGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGA ...(((((((..(((......((((..((((.......((((..(((......))).))))((....)).(((...(((......))).))).))))..))))......))).))))))) (-28.48 = -28.52 + 0.04)


| Location | 17,282,696 – 17,282,816 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282696 120 - 23771897 UCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCCCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCGAAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUU ..((((((..(((...(((((..((((.(((.(((......))).))).((..(((((....(((((...)))))...)))))..))..))))..)))))......)))))))))..... ( -25.10) >DroSec_CAF1 166421 120 - 1 UCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCGAAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUU ..((((((..(((...(((((..((((.(((.(((......)))...)))...(((((....(((((...)))))...)))))......))))..)))))......)))))))))..... ( -25.70) >DroSim_CAF1 162607 120 - 1 UCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCGAAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUU ..((((((..(((...(((((..((((.(((.(((......)))...)))...(((((....(((((...)))))...)))))......))))..)))))......)))))))))..... ( -25.70) >DroEre_CAF1 169463 120 - 1 UCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCGAAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUU ..((((((..(((...(((((..((((.(((.(((......)))...)))...(((((....(((((...)))))...)))))......))))..)))))......)))))))))..... ( -25.70) >DroYak_CAF1 174516 120 - 1 UCCAAUUGCCACUUCUGUGCAUCUGAGUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCGAAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUU ..((((((..(((...(((((..((((.(((.(((......)))...)))...(((((....(((((...)))))...)))))......))))..)))))......)))))))))..... ( -25.40) >consensus UCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCGAAAAAAGUUUUCAUUUGCAUGUUUUUAGUUAAUUGAAUUU ..((((((..(((...(((((..((((.(((.(((......))).))).((..(((((....(((((...)))))...)))))..))..))))..)))))......)))))))))..... (-25.36 = -25.20 + -0.16)



| Location | 17,282,736 – 17,282,856 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.64 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282736 120 + 23771897 CGAGGGCGACAAAGCCGCGGGGCGAAAGCAGGGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGAAGUGGCCACCAGGCGCAUCCUCUAUCCUCUAUCCUCCAGU .(((((((.......)))(((((....))((((...(((......))).........(((((.(....(((((.((.....)).)))))...).))))))))).))).....)))).... ( -36.60) >DroSec_CAF1 166461 113 + 1 CGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGAAGUGGCCACCAGGCGCAUCCUCU-------AUCCUCCAGU .(((((((.......)))(((((....)).(((...(((......))).))).....(((((.(....(((((.((.....)).)))))...).)))))))).-------..)))).... ( -35.40) >DroSim_CAF1 162647 113 + 1 CGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGAAGUGGCCACCAGGCGCAUCCUCU-------AUCCUCCAGU .(((((((.......)))(((((....)).(((...(((......))).))).....(((((.(....(((((.((.....)).)))))...).)))))))).-------..)))).... ( -35.40) >DroEre_CAF1 169503 111 + 1 CGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGAAGUGGCCACCAGGCGCCUCC--U-------AUUCCGCAUU ....(((......)))(((((((....)).......(((((...((((.((.....)).))))......)))))...(((.(((.((....))))).)))--.-------..)))))... ( -34.60) >DroYak_CAF1 174556 113 + 1 CGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCACUCAGAUGCACAGAAGUGGCAAUUGGAAGUGGCCACCAGGCGCAUCCUCU-------AUUCCCCAUU ....(((......)))..(((((....))((((...(((......))).........(((((.(....(((((.((.....)).)))))...).)))))))))-------....)))... ( -37.20) >consensus CGAGGGCGACAAAGCCGCGGGGCGAAAGCAGAGAAAUGCUAAAAUGCAACUCAUUCAGAUGCACAGAAGUGGCAAUUGGAAGUGGCCACCAGGCGCAUCCUCU_______AUCCUCCAGU ....(((......)))(.(((((....)).(((...(((......))).))).....(((((.(....(((((.((.....)).)))))...).))))).............))).)... (-32.08 = -31.64 + -0.44)

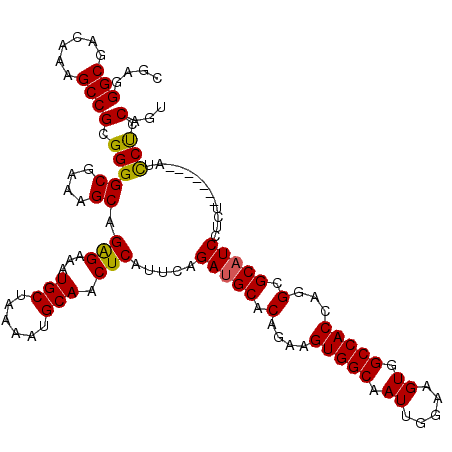

| Location | 17,282,736 – 17,282,856 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -33.44 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17282736 120 - 23771897 ACUGGAGGAUAGAGGAUAGAGGAUGCGCCUGGUGGCCACUUCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCCCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCG ....((((...(((..(((.((((((((..((((((...........))))))...))))))))....(((.(((......))).))).)))..))).....(((((...))))))))). ( -39.40) >DroSec_CAF1 166461 113 - 1 ACUGGAGGAU-------AGAGGAUGCGCCUGGUGGCCACUUCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCG ....((((..-------...((((((((..((((((...........))))))...))))))))(((.(((.(((......)))...)))....))).....(((((...))))))))). ( -38.10) >DroSim_CAF1 162647 113 - 1 ACUGGAGGAU-------AGAGGAUGCGCCUGGUGGCCACUUCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCG ....((((..-------...((((((((..((((((...........))))))...))))))))(((.(((.(((......)))...)))....))).....(((((...))))))))). ( -38.10) >DroEre_CAF1 169503 111 - 1 AAUGCGGAAU-------A--GGAGGCGCCUGGUGGCCACUUCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCG ...((((...-------.--(((((((((.((((((...........))))))...).))........(((.(((......)))...))).))))))...))))(((......))).... ( -34.50) >DroYak_CAF1 174556 113 - 1 AAUGGGGAAU-------AGAGGAUGCGCCUGGUGGCCACUUCCAAUUGCCACUUCUGUGCAUCUGAGUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCG ...((((...-------.((((((((((..((((((...........))))))...))))))))(((((((.(((......)))...))).)))))).))))(((((...)))))..... ( -41.90) >consensus ACUGGAGGAU_______AGAGGAUGCGCCUGGUGGCCACUUCCAAUUGCCACUUCUGUGCAUCUGAAUGAGUUGCAUUUUAGCAUUUCUCUGCUUUCGCCCCGCGGCUUUGUCGCCCUCG ....((((............((((((((..((((((...........))))))...))))))))((..(((((((.....((((......))))........)))))))..))..)))). (-33.44 = -34.00 + 0.56)

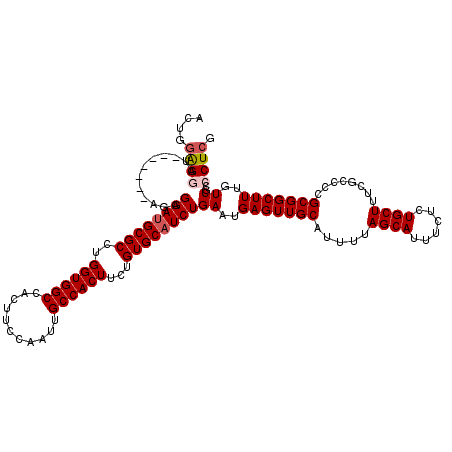
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:28 2006