
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,281,667 – 17,281,825 |
| Length | 158 |
| Max. P | 0.961072 |

| Location | 17,281,667 – 17,281,785 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -30.20 |
| Energy contribution | -31.20 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17281667 118 + 23771897 --GCAACAUGUUGCCGGGCAACCGGGCAUAAAUGAGGGGCUUAUCCUAUCGGGCAGCAAAACUUGAGCAAUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAA --....(((..((((((....)).))))...)))....((((((((...(((((((....((((..(((((((((....)))))).)))..)))))))).))).))))))))........ ( -41.90) >DroSec_CAF1 165354 117 + 1 --GCAACAUGUUGCCGGGCAAC-GGGCAUAAAUGAGGAACUUAUCCUAUCGAGCAGCACAACUUGAGCAUUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAA --....(((..((((.(....)-.))))...))).....(((((((...((....(((((((((..(((((((((....))))).))))..)))).))))))).)))))))......... ( -36.90) >DroSim_CAF1 161590 117 + 1 --GCAACAUGUUGCCGGGCAAC-GGGCAUAAAUGGGGGGCUUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGAAAAA --.(..(((..((((.(....)-.))))...)))..).((((((((...((....(((((((((..(((((((((....)))))).)))..)))).))))))).))))))))........ ( -42.60) >DroEre_CAF1 168372 117 + 1 --GCAACAUGUUGCCGGGCAAC-GGGCAUAAAUGAGGGGCUUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUACGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAA --....(((..((((.(....)-.))))...)))....((((((((...((....((((((((((((((((...)))))))..........)))).))))))).))))))))........ ( -37.50) >DroYak_CAF1 173356 119 + 1 CGGCAACAUGUUGCCGGGCAAC-GGGCAUAAAUGAGGGGCUUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUACGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAA ......(((..((((.(....)-.))))...)))....((((((((...((....((((((((((((((((...)))))))..........)))).))))))).))))))))........ ( -37.50) >consensus __GCAACAUGUUGCCGGGCAAC_GGGCAUAAAUGAGGGGCUUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAA ...(..(((..((((.........))))...)))..).((((((((...((....(((((((((..(((((((((....)))))).)))..)))).))))))).))))))))........ (-30.20 = -31.20 + 1.00)

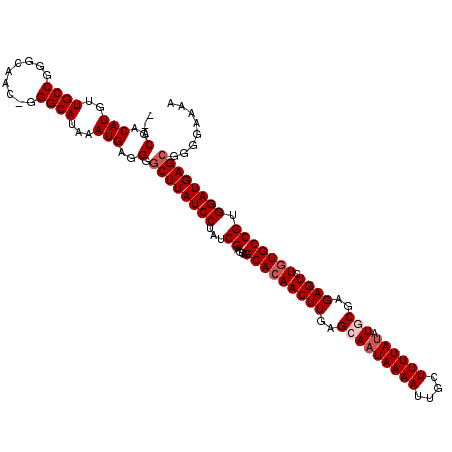

| Location | 17,281,705 – 17,281,825 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17281705 120 + 23771897 UUAUCCUAUCGGGCAGCAAAACUUGAGCAAUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAAUAGCCUGAAAUAUUCCUGGCAUGCUCAGAUAUACAACCAU ......((((((((.(((......(((((((...))))))).....)))........((((((.(((((..((((........))))...))))).)))))))))).))))......... ( -32.50) >DroSec_CAF1 165391 120 + 1 UUAUCCUAUCGAGCAGCACAACUUGAGCAUUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAAUAGCCUGAAAUAUUCCUGGCAUGCUCAGAUAUACAACCAU ......((((((((((((((((((..(((((((((....))))).))))..)))).))))).........((.(((((.((........)).))))).)).))))).))))......... ( -34.80) >DroSim_CAF1 161627 119 + 1 UUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGAAAAAUAGUC-GAAAUAUUUCUGACAUGCUCAGAUAUACAACCUU ......((((((((((((((((((..(((((((((....)))))).)))..)))).)))))(((......))).........(((-(((....))).))).))))).))))......... ( -30.10) >DroEre_CAF1 168409 120 + 1 UUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUACGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAAUAGCCUGAAAUAUUCCUGGCAUGCUCAGAUAUACAACCAU ......(((((((((((((((((((((((((...)))))))..........)))).))))).........((.(((((.((........)).))))).)).))))).))))......... ( -31.20) >DroYak_CAF1 173395 120 + 1 UUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUACGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAAUAGCCUGAAAUAUUCCUGGCAUGCUCAGAUAUACAACCAU ......(((((((((((((((((((((((((...)))))))..........)))).))))).........((.(((((.((........)).))))).)).))))).))))......... ( -31.20) >consensus UUAUCCUAUCGAGCAGCACAACUUGAGCAAUAAAAUUGCUUUUAUAUGCGAGAGUCUGUGCCGUGGAUGAGCGGGGAAAAUAGCCUGAAAUAUUCCUGGCAUGCUCAGAUAUACAACCAU ......((((((((((((((((((..(((((((((....)))))).)))..)))).))))).........((.(((((.((........)).))))).)).))))).))))......... (-28.94 = -29.46 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:22 2006