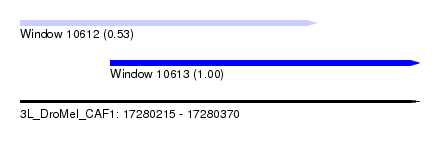
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,280,215 – 17,280,370 |
| Length | 155 |
| Max. P | 0.999768 |

| Location | 17,280,215 – 17,280,330 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -23.82 |
| Energy contribution | -23.66 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17280215 115 + 23771897 CAGGAUGAAAUAGCUUGCGCCGCGGA-----GCGAAAAUAAAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGC ..(((..(.((((((.......((((-----..((...(((((....))))).))..))))......))))))..)..)))....(((((((((((((.....)))....)))))))))) ( -27.92) >DroSec_CAF1 164015 115 + 1 CAGGAUGAAAAAGCUUGCGCCGCGGA-----GCGAAAAUAAAUAAAUAUUUGGUUAAUUCAAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGC ..(((..(....((((((...)).))-----)).................(((((((........)))))))...)..)))....(((((((((((((.....)))....)))))))))) ( -24.90) >DroSim_CAF1 160156 115 + 1 CAGGAUGAAAAAGCUUGCGCCGCGGA-----GCGAAAAUAAAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGC ..(((..(....((((((...)).))-----)).................(((((((........)))))))...)..)))....(((((((((((((.....)))....)))))))))) ( -24.90) >DroEre_CAF1 166953 115 + 1 CAGGAUGGAAAAGCUUGCGCCGCGAA-----GCGAAAAUAAAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGC ..(((..(....((((((...)).))-----)).................(((((((........)))))))...)..)))....(((((((((((((.....)))....)))))))))) ( -25.20) >DroYak_CAF1 171936 120 + 1 CAGGAUGAAAAAGCUUGCGCCGCGAAGCGAAGCGAAAAUAAAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGC ..(((..(....((((.(((......))))))).................(((((((........)))))))...)..)))....(((((((((((((.....)))....)))))))))) ( -29.20) >consensus CAGGAUGAAAAAGCUUGCGCCGCGGA_____GCGAAAAUAAAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGC ..(((..(............(((........)))................(((((((........)))))))...)..)))....(((((((((((((.....)))....)))))))))) (-23.82 = -23.66 + -0.16)

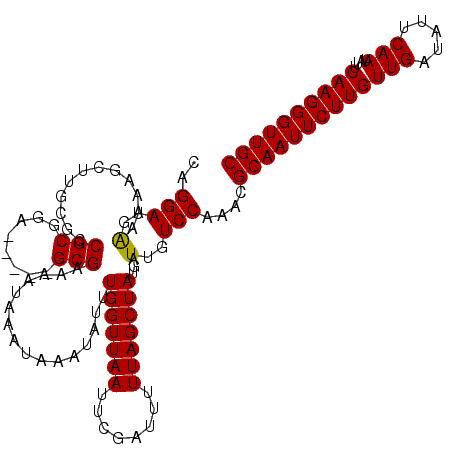
| Location | 17,280,250 – 17,280,370 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.26 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17280250 120 + 23771897 AAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGCGACUGGGCAUCGCGGCUCGAAAUAAGAAUACAACUCGGCA ...........((((.((((.(((((.(((..((((.(((((..((((((((((((((.....)))....)))))))))))..)))))))))..))).)))))..))))..))))..... ( -35.40) >DroSec_CAF1 164050 120 + 1 AAUAAAUAUUUGGUUAAUUCAAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGCGACUGGGCAUCGCGGCUCGAAAUAAGAAUACAACUCGGCA ...........((((.((((.(((((.(((..((((.(((((..((((((((((((((.....)))....)))))))))))..)))))))))..))).)))))..))))..))))..... ( -34.70) >DroSim_CAF1 160191 120 + 1 AAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGCGACUGGGCAUCGCGGCUCGAAAUAAGAAUACAACUCGGCA ...........((((.((((.(((((.(((..((((.(((((..((((((((((((((.....)))....)))))))))))..)))))))))..))).)))))..))))..))))..... ( -35.40) >DroEre_CAF1 166988 120 + 1 AAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGCGACUGGGCAUCGCGGCUCGAAAUAAGAAUACAACUCGGCA ...........((((.((((.(((((.(((..((((.(((((..((((((((((((((.....)))....)))))))))))..)))))))))..))).)))))..))))..))))..... ( -35.40) >DroYak_CAF1 171976 120 + 1 AAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGCGACUGGGCAUCGCGGCUCGAAAUAAGAAUACAACUCGGCA ...........((((.((((.(((((.(((..((((.(((((..((((((((((((((.....)))....)))))))))))..)))))))))..))).)))))..))))..))))..... ( -35.40) >consensus AAUAAAUAUUUGGUUAAUUCGAUUUUUAGCUAUGAUUGUCCAAACGCAAUUCUUGUUGAUAUUCAAUAUUCAAGGGUUGCGACUGGGCAUCGCGGCUCGAAAUAAGAAUACAACUCGGCA ...........((((.((((.(((((.(((..((((.(((((..((((((((((((((.....)))....)))))))))))..)))))))))..))).)))))..))))..))))..... (-35.26 = -35.26 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:17 2006