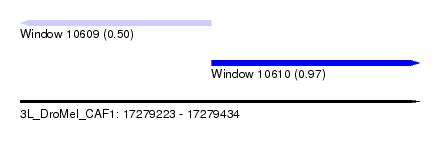
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,279,223 – 17,279,434 |
| Length | 211 |
| Max. P | 0.972708 |

| Location | 17,279,223 – 17,279,324 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -22.58 |
| Energy contribution | -24.89 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17279223 101 - 23771897 UUUCCUUGCCACUUUUCC-GGGGGUCU-----------CCACCAGCUCCUCCUUGGCUGCGCAAUUUCCAAGGAUCAGAUAGUCCUUCGUGUCGGGCAGCUGUUAUUACAAUA ..................-(((((.((-----------.....))..)))))..((((((.(.((..(.((((((......)))))).).)).).))))))............ ( -24.30) >DroSec_CAF1 162921 112 - 1 UUUCCUUGCCACUUUUCCUGGGGGUCUG-CCUCCAGCUCCUGCGGCUCCUCCUUGGCUGCGCAAUUUCCAAGGAUCAGAUAGUCCUUCGUGUCGGGCAGCUGUUAUUACAAUA .......(((.(.....(((((((....-))))))).....).)))........((((((.(.((..(.((((((......)))))).).)).).))))))............ ( -34.80) >DroSim_CAF1 157980 112 - 1 UUUCCUUGCCACUUUUCCUGGGGGUCUG-CCUCCAGCUCCUGCGGCUCCUCCUUGGCUGCGCAAUUUCCAAGGAUCAGAUAGUCCUUCGUGUCGGGCAGCUGUUAUUACAAUA .......(((.(.....(((((((....-))))))).....).)))........((((((.(.((..(.((((((......)))))).).)).).))))))............ ( -34.80) >DroEre_CAF1 165973 109 - 1 UUUCCUUGCCAUU-UUCCUGGGGGUC--UCCUCCAGCUCCUCUGGCUCCUCCUUGCCUGCGCA-UUUCCUAGGAUCAGAUAGUCCUUCGUGUCGGGCAGCUGUUAUUACAAUA .......((((..-...(((((((..--.)))))))......))))......(((((((.(((-(.....(((((......)))))..))))))))))).............. ( -31.20) >consensus UUUCCUUGCCACUUUUCCUGGGGGUCU__CCUCCAGCUCCUCCGGCUCCUCCUUGGCUGCGCAAUUUCCAAGGAUCAGAUAGUCCUUCGUGUCGGGCAGCUGUUAUUACAAUA .......(((.......(((((((.....))))))).......)))........((((((.(.....(.((((((......)))))).)....).))))))............ (-22.58 = -24.89 + 2.31)



| Location | 17,279,324 – 17,279,434 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -33.70 |
| Energy contribution | -35.08 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17279324 110 + 23771897 AGGCGAAGGAAACGUCGAGGAAACGGCCACGCAGCAAUGCAUUUUGCAGUUAGCAACAAUGGGCCAGCGGUCAAAGCCACAAUAAAAGCGUGGUGUGGCAUCGCUGGAUG .(((((.(....).))..(....).)))..(((....)))...((((.....)))).......(((((((((...(((((.........)))))...).))))))))... ( -38.30) >DroSec_CAF1 163033 99 + 1 AGGCCAA-----------GGAAACGGCCACGCAGCAAUGCAUUUUGCAGUUAGCAACAAUGGGCCAGCGGUCAAAGCCACAAUAAAAGCGUGGUGUGGCAUCGCUGGAUG .((((..-----------(....)))))..(((....)))...((((.....)))).......(((((((((...(((((.........)))))...).))))))))... ( -36.70) >DroSim_CAF1 158092 110 + 1 AGGCGAAGGAAACGCCGAGGAAACGGCCACGCAGCAAUGCAUUUUGCAGUUAGCAACAAUGGGCCAGCGGUCAAAGCCACAAUAAAAGCGUGGUGUGGCAUCGCUGGAUG .(((((.(....)((((.(....)((((..(((....)))...((((.....)))).....))))..))))....((((((.((......)).)))))).)))))..... ( -39.90) >DroEre_CAF1 166082 110 + 1 AGGCGAAGGAAACGCCGAGGAAACGGCCACGCAGCAAUGCAUUUUGCAGUUAGCAACAAUGGGCCAGCGGUCAAAGCCACAAUAAAAGCGUGGCGUGGCAUCGCUGGAUG .(((((.(....)((((.(....).(((((((.((..(.((((((((.....)))).)))).)...))(((....))).........))))))).)))).)))))..... ( -41.20) >consensus AGGCGAAGGAAACGCCGAGGAAACGGCCACGCAGCAAUGCAUUUUGCAGUUAGCAACAAUGGGCCAGCGGUCAAAGCCACAAUAAAAGCGUGGUGUGGCAUCGCUGGAUG .(((...(....).....(....).)))..(((....)))...((((.....)))).......(((((((((...(((((.........)))))...).))))))))... (-33.70 = -35.08 + 1.37)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:14 2006