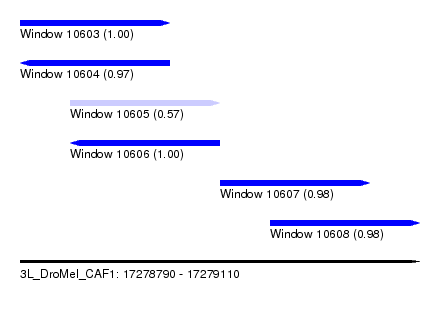
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,278,790 – 17,279,110 |
| Length | 320 |
| Max. P | 0.999574 |

| Location | 17,278,790 – 17,278,910 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -22.36 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17278790 120 + 23771897 GAGUGUAUAAAAAUGUCGCUCAAUUAAAGUGUAGACACUUAAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCU (((((((..........((((.....((((((.((..........)).))))))......(.((((.......)))))))))(((((((...)))))))....))))))).......... ( -26.30) >DroSec_CAF1 162488 120 + 1 GAGUGUAUAAAAAUGUCGCUCAAUUAAAGUGUAGACACUUAAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCU (((((((..........((((.....((((((.((..........)).))))))......(.((((.......)))))))))(((((((...)))))))....))))))).......... ( -26.30) >DroSim_CAF1 157547 120 + 1 GAGUGUAUAAAAAUGUCGCUCAAUUAAAGUGUAGACACUUAAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCU (((((((..........((((.....((((((.((..........)).))))))......(.((((.......)))))))))(((((((...)))))))....))))))).......... ( -26.30) >DroEre_CAF1 165541 120 + 1 CAGUGUAUAAAAAUGUCGCUCAAUUAAAGUGUAGACACUUAAAAUUCAACACUUAAUUUUGUGCGUCACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCCCAACAAAGUCU ..(((((..........((((.((((..((((.(((.(.(((((((........))))))).).)))))))))))...))))(((((((...)))))))....)))))............ ( -21.60) >DroYak_CAF1 170444 120 + 1 GAGAGUAUAAAAAUGUCGCUCAAUUAAAGUGUAGACACUUAAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCCCAACAAAGUCU ....(((..........((((.....((((((.((..........)).))))))......(.((((.......)))))))))(((((((...)))))))....))).............. ( -18.10) >consensus GAGUGUAUAAAAAUGUCGCUCAAUUAAAGUGUAGACACUUAAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCU (((((((..........((((.....((((((.((..........)).))))))......(.((((.......)))))))))(((((((...)))))))....))))))).......... (-22.36 = -23.16 + 0.80)

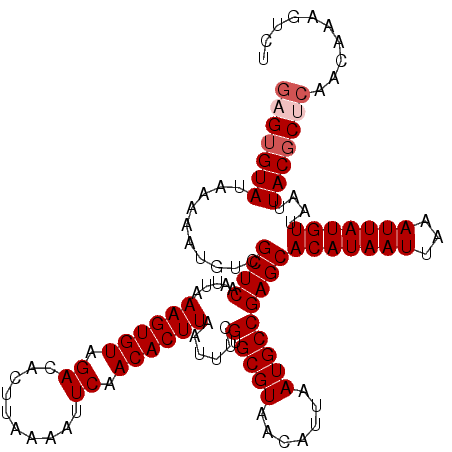
| Location | 17,278,790 – 17,278,910 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -26.20 |
| Energy contribution | -25.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17278790 120 - 23771897 AGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUUAAGUGUCUACACUUUAAUUGAGCGACAUUUUUAUACACUC ......((((((((((((((((......))))))))..))))))))..(((((..(((((........))))(.((((..(((((....))))).)))).))..)))))........... ( -26.30) >DroSec_CAF1 162488 120 - 1 AGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUUAAGUGUCUACACUUUAAUUGAGCGACAUUUUUAUACACUC ......((((((((((((((((......))))))))..))))))))..(((((..(((((........))))(.((((..(((((....))))).)))).))..)))))........... ( -26.30) >DroSim_CAF1 157547 120 - 1 AGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUUAAGUGUCUACACUUUAAUUGAGCGACAUUUUUAUACACUC ......((((((((((((((((......))))))))..))))))))..(((((..(((((........))))(.((((..(((((....))))).)))).))..)))))........... ( -26.30) >DroEre_CAF1 165541 120 - 1 AGACUUUGUUGGGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUGACGCACAAAAUUAAGUGUUGAAUUUUAAGUGUCUACACUUUAAUUGAGCGACAUUUUUAUACACUG ......((((((((((((((((......))))))))..))))))))..(((((..(((.(((....((((((.((..........)).))))))...))).))))))))........... ( -25.50) >DroYak_CAF1 170444 120 - 1 AGACUUUGUUGGGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUUAAGUGUCUACACUUUAAUUGAGCGACAUUUUUAUACUCUC ......((((((((((((((((......))))))))..))))))))..(((((..(((((........))))(.((((..(((((....))))).)))).))..)))))........... ( -25.40) >consensus AGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUUAAGUGUCUACACUUUAAUUGAGCGACAUUUUUAUACACUC ......((((((((((((((((......))))))))..))))))))..(((((..(((.(((....((((((.((..........)).))))))...))).))))))))........... (-26.20 = -25.80 + -0.40)

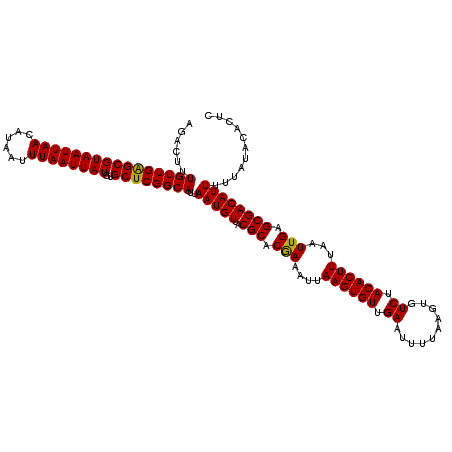
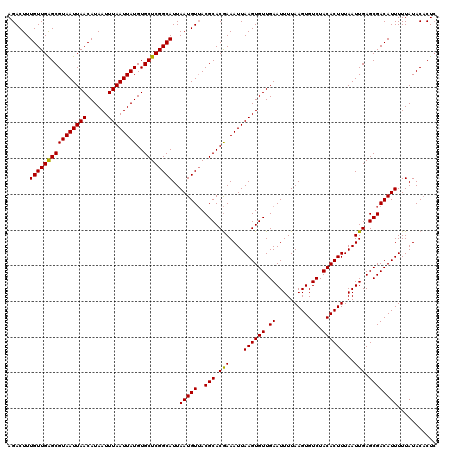
| Location | 17,278,830 – 17,278,950 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17278830 120 + 23771897 AAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCUAUUAACAACACACUGCUAGAACGAGAUAGACAGCUGAAAU ...............((((((.((..............((((...(((((((......))))))).))))......(((((((.....................))))))).)))))))) ( -18.80) >DroSec_CAF1 162528 120 + 1 AAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCUAUUAACAACACACUGCUAGAACGAGAUAGACAGCUGAAAU ...............((((((.((..............((((...(((((((......))))))).))))......(((((((.....................))))))).)))))))) ( -18.80) >DroSim_CAF1 157587 120 + 1 AAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCUAUUAACAACACACUGCUAGAACGAGAUAGACAGCUGAAAU ...............((((((.((..............((((...(((((((......))))))).))))......(((((((.....................))))))).)))))))) ( -18.80) >DroEre_CAF1 165581 120 + 1 AAAAUUCAACACUUAAUUUUGUGCGUCACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCCCAACAAAGUCUAUUAACAACACACUGCUAGAACGAGAUAGACAGUUGAAAU ....((((((..((((((.(((((.((.((....))..))))))).))))))........................(((((((.....................))))))).)))))).. ( -21.50) >DroYak_CAF1 170484 120 + 1 AAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCCCAACAAAGUCUAUUAACAACACACUGCAGGAACGAGAUAGACAGUUGAGCU ....((((((..........(.((((((.......((...))(((((((...)))))))...)))))).)......(((((((.......(......)......))))))).)))))).. ( -20.52) >consensus AAAAUUCAACACUUAAUUUCGUGCGUAACAUUAAUGCCGAGCACAUAAUUAAAUUAUGUUAAUUACGCUCAACAAAGUCUAUUAACAACACACUGCUAGAACGAGAUAGACAGCUGAAAU ....((((.(..........(.((((((.......((...))(((((((...)))))))...)))))).)......(((((((.....................))))))).).)))).. (-16.72 = -16.76 + 0.04)

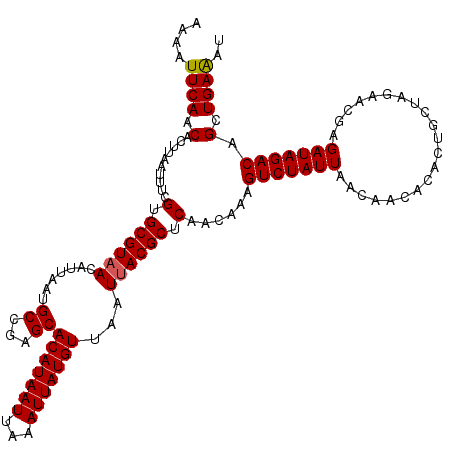
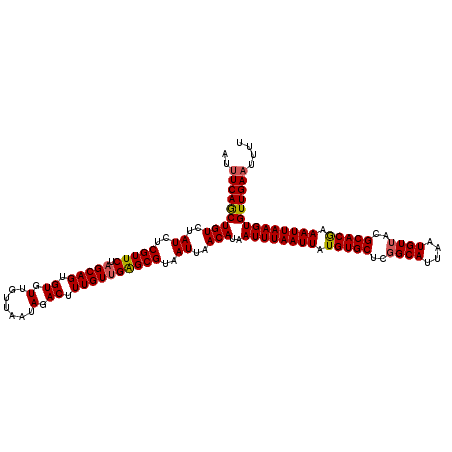
| Location | 17,278,830 – 17,278,950 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.40 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17278830 120 - 23771897 AUUUCAGCUGUCUAUCUCGUUCUAGCAGUGUGUUGUUAAUAGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUU ..(((((((((..((..(((((.(((((.((.(.......).)).))))))))))..))..)))..((((((((.(((((..((((....))))..))))).)))))))))))))).... ( -29.90) >DroSec_CAF1 162528 120 - 1 AUUUCAGCUGUCUAUCUCGUUCUAGCAGUGUGUUGUUAAUAGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUU ..(((((((((..((..(((((.(((((.((.(.......).)).))))))))))..))..)))..((((((((.(((((..((((....))))..))))).)))))))))))))).... ( -29.90) >DroSim_CAF1 157587 120 - 1 AUUUCAGCUGUCUAUCUCGUUCUAGCAGUGUGUUGUUAAUAGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUU ..(((((((((..((..(((((.(((((.((.(.......).)).))))))))))..))..)))..((((((((.(((((..((((....))))..))))).)))))))))))))).... ( -29.90) >DroEre_CAF1 165581 120 - 1 AUUUCAACUGUCUAUCUCGUUCUAGCAGUGUGUUGUUAAUAGACUUUGUUGGGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUGACGCACAAAAUUAAGUGUUGAAUUUU ..(((((((((..((..(((.(((((((.((.(.......).)).))))))))))..))..)))..((((((((.((((((((.((....))))).))))).)))))))))))))).... ( -30.00) >DroYak_CAF1 170484 120 - 1 AGCUCAACUGUCUAUCUCGUUCCUGCAGUGUGUUGUUAAUAGACUUUGUUGGGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUU .(((((((.((((((..((...(......)...))...))))))...)))))))....(((((((...((((((.(((((..((((....))))..))))).)))))))))))))..... ( -31.10) >consensus AUUUCAGCUGUCUAUCUCGUUCUAGCAGUGUGUUGUUAAUAGACUUUGUUGAGCGUAAUUAACAUAAUUUAAUUAUGUGCUCGGCAUUAAUGUUACGCACGAAAUUAAGUGUUGAAUUUU ..(((((((((..((..(((((.(((((.((.(.......).)).))))))))))..))..)))..((((((((.(((((..((((....))))..))))).)))))))))))))).... (-28.44 = -28.40 + -0.04)

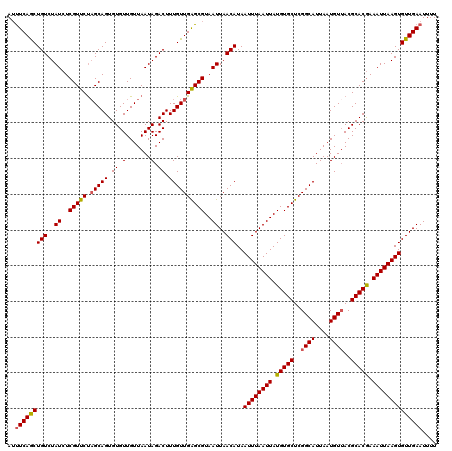
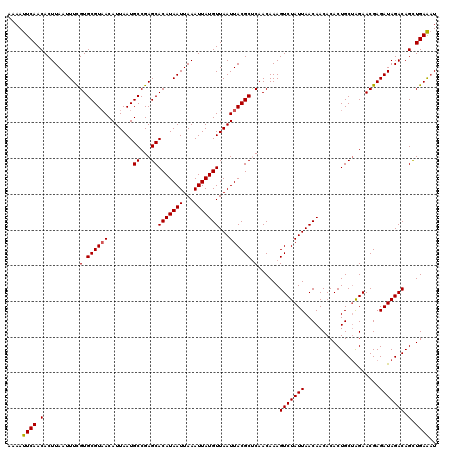
| Location | 17,278,950 – 17,279,070 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -27.82 |
| Energy contribution | -27.46 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17278950 120 + 23771897 GGCUGACAGAAGGAGACGGACACUGAGGCGAAAUCCCUUUUGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUC (..((((((((((.((((..(.....).))...)))))))))((((.....))))................)))..)...(((((((..(((((.......))))).....))))))).. ( -29.40) >DroSec_CAF1 162648 120 + 1 GGCUGACAGAAGGAGACGGACACCGGGGCCAAAUCCCUUUUGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUC (..((((((((((.((.((.(......)))...)))))))))((((.....))))................)))..)...(((((((..(((((.......))))).....))))))).. ( -31.10) >DroSim_CAF1 157707 120 + 1 GGCUGACAGAAGGAGACGGACACCGGGGCCAAAUCCCUUUUGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUC (..((((((((((.((.((.(......)))...)))))))))((((.....))))................)))..)...(((((((..(((((.......))))).....))))))).. ( -31.10) >DroEre_CAF1 165701 120 + 1 GCCUGAUAGAAGGAGACGGACACUGAGGCUAAAUCCCUUUUGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUC ...((((.(((((.......((..((((.......)))).))((((.....)))).............))))).))))..(((((((..(((((.......))))).....))))))).. ( -29.50) >DroYak_CAF1 170604 120 + 1 GCCUGAUGGAAGGAGACGGACGCAGAAGCUAAAUCCCUUUUGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUC ...((((.(((((.........((((((........))))))((((.....)))).............))))).))))..(((((((..(((((.......))))).....))))))).. ( -30.50) >consensus GGCUGACAGAAGGAGACGGACACCGAGGCCAAAUCCCUUUUGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUC (..((((((((((.((.((.(......)))...)))))))))((((.....))))................)))..)...(((((((..(((((.......))))).....))))))).. (-27.82 = -27.46 + -0.36)

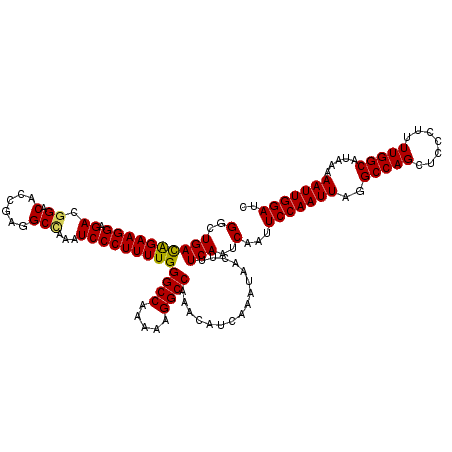
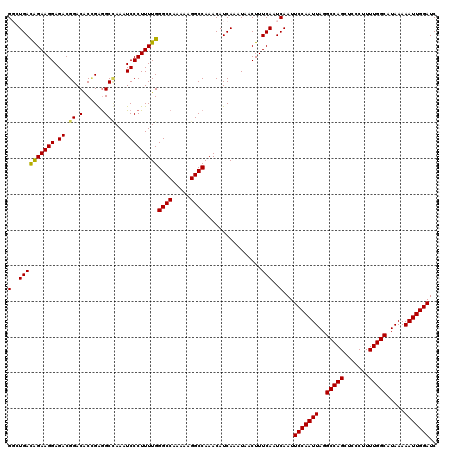
| Location | 17,278,990 – 17,279,110 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17278990 120 + 23771897 UGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUCCAGAACGGAACGACAAUUUAAUUUCAAGUUUAAUGGCAAA ..((((.....)))).....(((...(((((.........(((((((..(((((.......))))).....)))))))(((.....)))................)))))...))).... ( -23.80) >DroSec_CAF1 162688 120 + 1 UGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUCCAGAACGGAACGACAAUUUAAUUUCAAGUUUAAUGGCAAA ..((((.....)))).....(((...(((((.........(((((((..(((((.......))))).....)))))))(((.....)))................)))))...))).... ( -23.80) >DroSim_CAF1 157747 120 + 1 UGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUCCAGAACGGAACGACAAUUUAAUUUCAAGUUUAAUGGCAAA ..((((.....)))).....(((...(((((.........(((((((..(((((.......))))).....)))))))(((.....)))................)))))...))).... ( -23.80) >DroEre_CAF1 165741 120 + 1 UGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUCCAGAACGGAACGACAAUUUAAUUUCAAGUUUAAUGGCAAA ..((((.....)))).....(((...(((((.........(((((((..(((((.......))))).....)))))))(((.....)))................)))))...))).... ( -23.80) >DroYak_CAF1 170644 120 + 1 UGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUCCAGAACGGAACGACAAUUUAAUUUCAAGUUUAAUGGCAAA ..((((.....)))).....(((...(((((.........(((((((..(((((.......))))).....)))))))(((.....)))................)))))...))).... ( -23.80) >consensus UGGGCCAAAAAGGCCAAACAUCAAAUAACUUUCAAUCAAUUCCAAUUAGGCCAGCUCCCUUUUGGCAUAAAAAUUGGAUCCAGAACGGAACGACAAUUUAAUUUCAAGUUUAAUGGCAAA ..((((.....)))).....(((...(((((.........(((((((..(((((.......))))).....)))))))(((.....)))................)))))...))).... (-23.80 = -23.80 + 0.00)

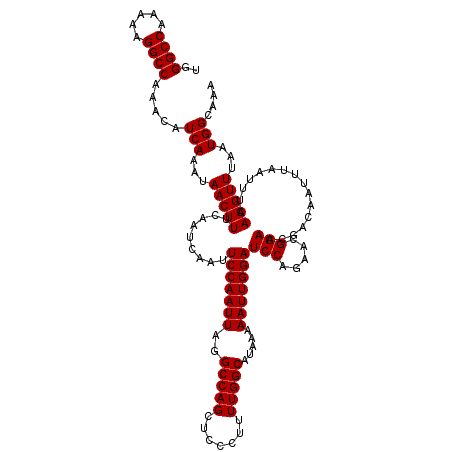
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:13 2006