
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,276,788 – 17,276,904 |
| Length | 116 |
| Max. P | 0.972015 |

| Location | 17,276,788 – 17,276,904 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -34.86 |
| Energy contribution | -35.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
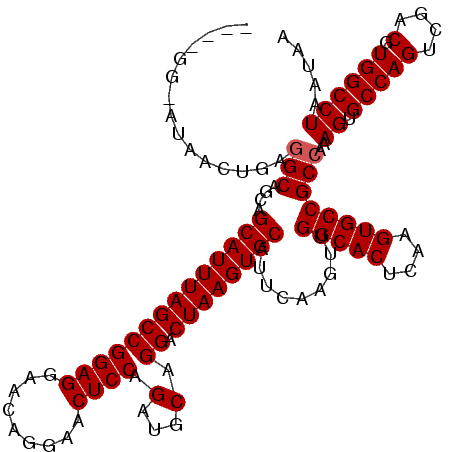
>3L_DroMel_CAF1 17276788 116 + 23771897 ----GAAAUAACUGAGGCAGCAGCAUUUAGCCGGAGGAACAGGAACUCCAGAUCCAGGACUAAGUGCAUUCAAGUUGGCACUCAAGUGCCGCCAAAGUGCCAGUCGACGUGGCCUAAUAA ----......(((..(((....((((((((((...(((.(.((....)).).))).)).)))))))).........(((((....))))))))..)))(((((....).))))....... ( -39.20) >DroSec_CAF1 160161 115 + 1 ----GG-AUAACUGAGGCAGCAGCAUUUAGCCGGAGGAACAGGAACUCCAGAUGCAGGACUAAGUGCAUUCAAGUUGGCACUCAAGUGCCGCCAAAGUGCCAGUCGACGUGGCCUAAUAA ----..-...(((..(((....((((((((((((((.........)))).(...).)).)))))))).........(((((....))))))))..)))(((((....).))))....... ( -36.80) >DroSim_CAF1 155490 115 + 1 ----GG-AUAACUGAGGCAGCAGCAUUUAGCCGGAGGAACAGGAACUCCAGAUGCAGGACUAAGUGCAUUCAAGUUGGCACUCAAGUGCCGCCAAAGUGCCAGUCGACGUGGCCUAAUAA ----..-...(((..(((....((((((((((((((.........)))).(...).)).)))))))).........(((((....))))))))..)))(((((....).))))....... ( -36.80) >DroEre_CAF1 163147 119 + 1 ACUGGG-ACAUCUGAGGCAGCAGCAUUUAGCCGGAGGAACAGGAACUCCAGAUGCAGGACUAAGUGCAUUCAAGUUGGCACUCAAGUGCCGCCAAAGUGCCAGUCGACGUGGCCUAAUAA (((((.-((......(((....((((((((((((((.........)))).(...).)).)))))))).........(((((....))))))))...)).)))))................ ( -37.40) >DroYak_CAF1 168123 119 + 1 AGUGGG-ACAACUGAAGCAGCAGCAUUUAGCCGGAGGAACAGGAACUCCAGAUGCAGGACUAAGUGCAUUCAAGUUGGCACUCAAGUGCCGCCAAAGUGCCAGUCGACGUGGCCUAAUAA ..((((-.((((((..((....((((((((((((((.........)))).(...).)).))))))))......(.((((((....)))))).)...))..)))).....)).)))).... ( -34.20) >consensus ____GG_AUAACUGAGGCAGCAGCAUUUAGCCGGAGGAACAGGAACUCCAGAUGCAGGACUAAGUGCAUUCAAGUUGGCACUCAAGUGCCGCCAAAGUGCCAGUCGACGUGGCCUAAUAA ...............(((....((((((((((((((.........)))).(...).)).)))))))).........(((((....))))))))..((.(((((....).))))))..... (-34.86 = -35.06 + 0.20)

| Location | 17,276,788 – 17,276,904 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -33.86 |
| Energy contribution | -34.10 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276788 116 - 23771897 UUAUUAGGCCACGUCGACUGGCACUUUGGCGGCACUUGAGUGCCAACUUGAAUGCACUUAGUCCUGGAUCUGGAGUUCCUGUUCCUCCGGCUAAAUGCUGCUGCCUCAGUUAUUUC---- .......((((.(....)))))(((..(((((((.(..(((....)))..)..(((.((((((..(((...((....))...)))...)))))).))))))))))..)))......---- ( -38.60) >DroSec_CAF1 160161 115 - 1 UUAUUAGGCCACGUCGACUGGCACUUUGGCGGCACUUGAGUGCCAACUUGAAUGCACUUAGUCCUGCAUCUGGAGUUCCUGUUCCUCCGGCUAAAUGCUGCUGCCUCAGUUAU-CC---- .......((((.(....)))))(((..(((((((.((((((((..........))))))))....((((((((((.........))))))....)))))))))))..)))...-..---- ( -36.10) >DroSim_CAF1 155490 115 - 1 UUAUUAGGCCACGUCGACUGGCACUUUGGCGGCACUUGAGUGCCAACUUGAAUGCACUUAGUCCUGCAUCUGGAGUUCCUGUUCCUCCGGCUAAAUGCUGCUGCCUCAGUUAU-CC---- .......((((.(....)))))(((..(((((((.((((((((..........))))))))....((((((((((.........))))))....)))))))))))..)))...-..---- ( -36.10) >DroEre_CAF1 163147 119 - 1 UUAUUAGGCCACGUCGACUGGCACUUUGGCGGCACUUGAGUGCCAACUUGAAUGCACUUAGUCCUGCAUCUGGAGUUCCUGUUCCUCCGGCUAAAUGCUGCUGCCUCAGAUGU-CCCAGU ......(((...))).(((((((((..(((((((.((((((((..........))))))))....((((((((((.........))))))....)))))))))))..)).)).-.))))) ( -36.40) >DroYak_CAF1 168123 119 - 1 UUAUUAGGCCACGUCGACUGGCACUUUGGCGGCACUUGAGUGCCAACUUGAAUGCACUUAGUCCUGCAUCUGGAGUUCCUGUUCCUCCGGCUAAAUGCUGCUGCUUCAGUUGU-CCCACU ......((....(.(((((((......(((((((.((((((((..........))))))))....((((((((((.........))))))....)))))))))))))))))).-)))... ( -34.30) >consensus UUAUUAGGCCACGUCGACUGGCACUUUGGCGGCACUUGAGUGCCAACUUGAAUGCACUUAGUCCUGCAUCUGGAGUUCCUGUUCCUCCGGCUAAAUGCUGCUGCCUCAGUUAU_CC____ .......((((.(....)))))(((..(((((((.((((((((..........))))))))....((((((((((.........))))))....)))))))))))..))).......... (-33.86 = -34.10 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:05 2006