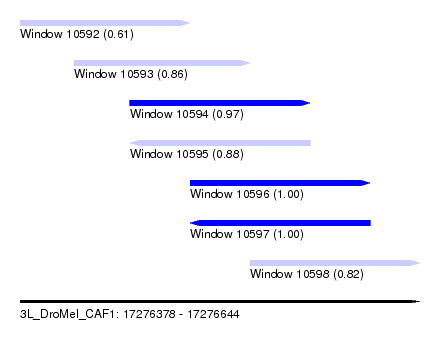
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,276,378 – 17,276,644 |
| Length | 266 |
| Max. P | 0.999795 |

| Location | 17,276,378 – 17,276,491 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -38.94 |
| Consensus MFE | -32.02 |
| Energy contribution | -31.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276378 113 + 23771897 AGAAAGUCGGGCAGUUUUCCGGGCUCCUUUGAAAGA-------AAAUGGAAAGGCGGUGAAGGCAGUCGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUC .......((((......))))(((((((((....))-------)........(((((.((..((((((((((((.(((.......)))..))))))))))))..)))))))..)))))). ( -40.20) >DroSec_CAF1 159734 112 + 1 AGA-AGUCGGGCAGUUUUCCGGGCUCCUUUGAGGGA-------AAAGGGGAAGGCGGUGAAGGCAGUGGAGCAGGAGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUU ...-(((((((((((((..((..(((((((......-------)))))))..((((((....((......))....))))))..)).)))))))))))))((..((((....)))))).. ( -37.70) >DroSim_CAF1 155062 113 + 1 AGAAAGUCGGGCAGUUUUCCGGGCUCCUUUGAGGGA-------AAAGGGGAAGGCGGUGAAGGCAGUGGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUU ....(((((((((((((..(...(((((((......-------)))))))..)(((..((.((((.(......).)))).))..))))))))))))))))((..((((....)))))).. ( -37.80) >DroEre_CAF1 162732 113 + 1 AGAAACCCGGGCAGUUUUCCGGGUUCCUUUGAAGGA-------AAAGGGAAAGGCGGAAAAGGCAGCGGACCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUC .....((..((((((((((((..(((((((......-------.)))))))...))))))).(((((.((.(((.(((.......)))..))).)).)))))....)))))..))..... ( -38.20) >DroYak_CAF1 167699 120 + 1 AGAAACUCGGGCAGUUUUCCAGGUUCCUUUGAAGGAAAUGGAAAAAGGGAAAGGCGGCGAAGGCGGUGGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUC ........((((..(((((((..(((((....))))).))))))).......(((((.((..(((((.((((((.(((.......)))..)))))).)))))..))))))).....)))) ( -40.80) >consensus AGAAAGUCGGGCAGUUUUCCGGGCUCCUUUGAAGGA_______AAAGGGAAAGGCGGUGAAGGCAGUGGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUC ......(((((......)))))(((((((.......................(((((.((..(((((.((((((....((....))....)))))).)))))..)))))))))))))).. (-32.02 = -31.90 + -0.12)

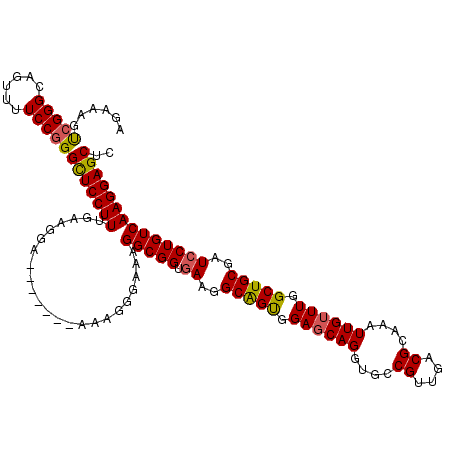

| Location | 17,276,414 – 17,276,531 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -32.92 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276414 117 + 23771897 ---AAAUGGAAAGGCGGUGAAGGCAGUCGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAU ---...(((...((((((....((((((((((((.(((.......)))..))))))))))))..((((....))))((((...(....)...)))))).)))).....)))......... ( -36.40) >DroSec_CAF1 159769 117 + 1 ---AAAGGGGAAGGCGGUGAAGGCAGUGGAGCAGGAGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUUCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAU ---...(((...((((((....((......))....))))))(.(((..((((((((.(((...((((....))))....))).))))))))..))))...)))................ ( -37.50) >DroSim_CAF1 155098 117 + 1 ---AAAGGGGAAGGCGGUGAAGGCAGUGGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUUCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAU ---....(((...(((((.((((((.(......).)))).)).))((..((((((((.(((...((((....))))....))).))))))))..)).))).)))................ ( -37.50) >DroEre_CAF1 162768 117 + 1 ---AAAGGGAAAGGCGGAAAAGGCAGCGGACCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAAGAGCGCGCGCCCAAUGUCAACUAAAUAU ---...(((....(((.....((((.(......).)))).....(((...(((((((.(((.((((((....))))..))))).)))))))...)))))).)))................ ( -36.40) >DroYak_CAF1 167739 120 + 1 GAAAAAGGGAAAGGCGGCGAAGGCGGUGGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAAGAGCGCGCGCCCAAUGUCAACUAAAUAU ((....(((...(((((((...((......))...)))))))(.(((...(((((((.(((.((((((....))))..))))).)))))))...))))...)))....)).......... ( -38.50) >consensus ___AAAGGGAAAGGCGGUGAAGGCAGUGGAGCAGGUGCCGUUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAU ......(((...(((((((...((......))...)))))))(.(((..((((((((.(((...((((....))))....))).))))))))..))))...)))................ (-32.92 = -33.88 + 0.96)

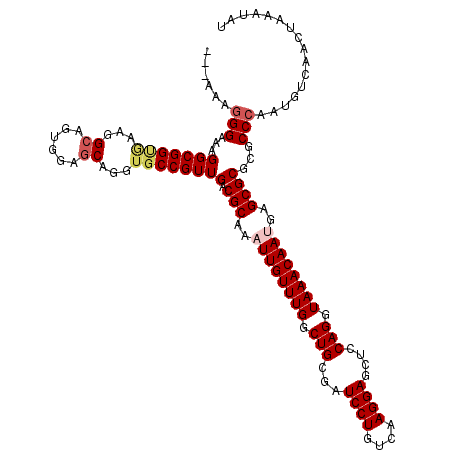
| Location | 17,276,451 – 17,276,571 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -39.44 |
| Energy contribution | -39.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276451 120 + 23771897 UUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGUCAGGAA (((((((..((((((((.(((.((((((....))))..))))).))))))))..))....(((((...(((.((......((..(....)..)).....)).)))..))))))))))... ( -40.30) >DroSec_CAF1 159806 120 + 1 UUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUUCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAA ..(.(((..((((((((.(((...((((....))))....))).))))))))..))))(((((((...(((.((......((..(....)..)).....)).)))..)))))))...... ( -41.50) >DroSim_CAF1 155135 120 + 1 UUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUUCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAA ..(.(((..((((((((.(((...((((....))))....))).))))))))..))))(((((((...(((.((......((..(....)..)).....)).)))..)))))))...... ( -41.50) >DroEre_CAF1 162805 120 + 1 UUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAAGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAA ..(.(((...(((((((.(((.((((((....))))..))))).)))))))...))))(((((((...(((.((......((..(....)..)).....)).)))..)))))))...... ( -39.10) >DroYak_CAF1 167779 120 + 1 UUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAAGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAA ..(.(((...(((((((.(((.((((((....))))..))))).)))))))...))))(((((((...(((.((......((..(....)..)).....)).)))..)))))))...... ( -39.10) >consensus UUGACGCAAAUUGUUUGGCUGCGAUCCUGUCAAGGAGCUCCAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAA ..(.(((..((((((((.(((...((((....))))....))).))))))))..))))(((((((...(((.((......((..(....)..)).....)).)))..)))))))...... (-39.44 = -39.68 + 0.24)

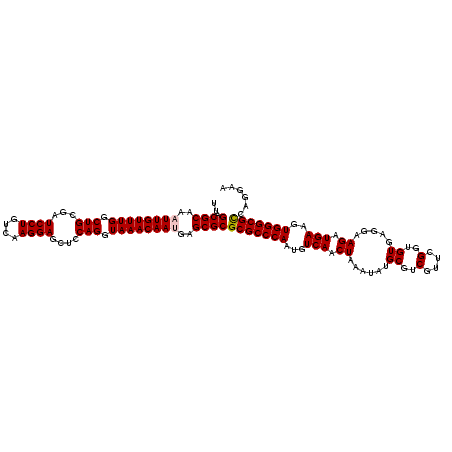
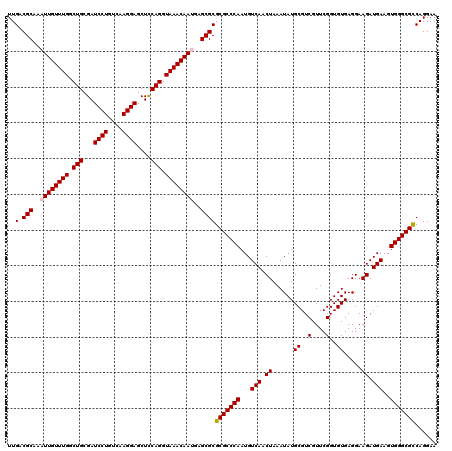
| Location | 17,276,451 – 17,276,571 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276451 120 - 23771897 UUCCUGACGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUGGAGCUCCUUGACAGGAUCGCAGCCAAACAAUUUGCGUCAA .....(.((((((...........((.....)).((((.........))))....)))))))((((..(((((((..(((((..((((....)))))).)))..)))))))..))))... ( -30.20) >DroSec_CAF1 159806 120 - 1 UUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUGAAGCUCCUUGACAGGAUCGCAGCCAAACAAUUUGCGUCAA ......(((((((...........((.....)).((((.........))))....)))))))((((..(((((((..(((....((((....))))...)))..)))))))..))))... ( -33.60) >DroSim_CAF1 155135 120 - 1 UUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUGAAGCUCCUUGACAGGAUCGCAGCCAAACAAUUUGCGUCAA ......(((((((...........((.....)).((((.........))))....)))))))((((..(((((((..(((....((((....))))...)))..)))))))..))))... ( -33.60) >DroEre_CAF1 162805 120 - 1 UUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCUUUGUUUACCUGGAGCUCCUUGACAGGAUCGCAGCCAAACAAUUUGCGUCAA .((((((((((((...........((.....)).((((.........))))....)))))))..(((((..(....)..))))).......))))).(((((.........))))).... ( -33.20) >DroYak_CAF1 167779 120 - 1 UUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCUUUGUUUACCUGGAGCUCCUUGACAGGAUCGCAGCCAAACAAUUUGCGUCAA .((((((((((((...........((.....)).((((.........))))....)))))))..(((((..(....)..))))).......))))).(((((.........))))).... ( -33.20) >consensus UUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUGGAGCUCCUUGACAGGAUCGCAGCCAAACAAUUUGCGUCAA ......(((((((...........((.....)).((((.........))))....)))))))((((...((((((..(((..(.((((....)))).).)))..))))))...))))... (-30.70 = -30.90 + 0.20)



| Location | 17,276,491 – 17,276,611 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -40.00 |
| Energy contribution | -39.84 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276491 120 + 23771897 CAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGUCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAA ...........(((((((..(((((...(((.((......((..(....)..)).....)).)))..)))))((((((....)))))).........)))))))................ ( -38.60) >DroSec_CAF1 159846 120 + 1 CAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAA ...........(((((((..(((((...(((.((......((..(....)..)).....)).)))..)))))((((((....)))))).........)))))))................ ( -41.30) >DroSim_CAF1 155175 120 + 1 CAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAA ...........(((((((..(((((...(((.((......((..(....)..)).....)).)))..)))))((((((....)))))).........)))))))................ ( -41.30) >DroEre_CAF1 162845 120 + 1 CAGGUAAACAAAGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAA ............((((((..(((((...(((.((......((..(....)..)).....)).)))..)))))((((((....)))))).........))))))................. ( -40.50) >DroYak_CAF1 167819 120 + 1 CAGGUAAACAAAGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAA ............((((((..(((((...(((.((......((..(....)..)).....)).)))..)))))((((((....)))))).........))))))................. ( -40.50) >consensus CAGGUAAACAAUGAGCGCGCGCCCAAUGUCAACUAAAUAUGCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAA ............((((((..(((((...(((.((......((..(....)..)).....)).)))..)))))((((((....)))))).........))))))................. (-40.00 = -39.84 + -0.16)



| Location | 17,276,491 – 17,276,611 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -33.66 |
| Energy contribution | -33.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276491 120 - 23771897 UUUCCUUUAUUUUAUUUGAGCGCUUAACUUUUGCCAGGACUUCCUGACGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUG ................(((((((...........((((....)))).((((((...........((.....)).((((.........))))....)))))).)))))))........... ( -30.40) >DroSec_CAF1 159846 120 - 1 UUUCCUUUAUUUUAUUUGAGCGCUUAACUUUUGCCAGGACUUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUG ................(((((((.........((((((....))))))(((((...........((.....)).((((.........))))....)))))..)))))))........... ( -36.10) >DroSim_CAF1 155175 120 - 1 UUUCCUUUAUUUUAUUUGAGCGCUUAACUUUUGCCAGGACUUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUG ................(((((((.........((((((....))))))(((((...........((.....)).((((.........))))....)))))..)))))))........... ( -36.10) >DroEre_CAF1 162845 120 - 1 UUUCCUUUAUUUUAUUUGAGCGCUUAACUUUUGCCAGGACUUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCUUUGUUUACCUG .................((((((.........((((((....))))))(((((...........((.....)).((((.........))))....)))))..))))))............ ( -34.90) >DroYak_CAF1 167819 120 - 1 UUUCCUUUAUUUUAUUUGAGCGCUUAACUUUUGCCAGGACUUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCUUUGUUUACCUG .................((((((.........((((((....))))))(((((...........((.....)).((((.........))))....)))))..))))))............ ( -34.90) >consensus UUUCCUUUAUUUUAUUUGAGCGCUUAACUUUUGCCAGGACUUCCUGGCGCCCACUUCAUCUUCCUCACACCGAACGACGCAUAUUUAGUUGACAUUGGGCGCGCGCUCAUUGUUUACCUG .................((((((.........((((((....))))))(((((...........((.....)).((((.........))))....)))))..))))))............ (-33.66 = -33.86 + 0.20)

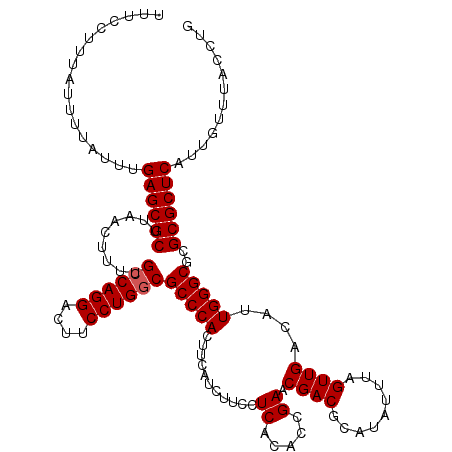
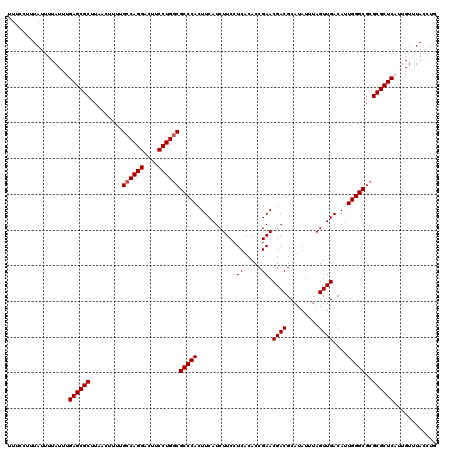
| Location | 17,276,531 – 17,276,644 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -23.18 |
| Energy contribution | -22.78 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276531 113 + 23771897 GCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGUCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAAAUGGCAAAAAGUUAGAUAGUGCGGGGAG---GGCUC---- .((((.(((.......))).))))....((((((((((....)))))).........(((((...................((((.....))))...)))))......---.))))---- ( -23.15) >DroSec_CAF1 159886 120 + 1 GCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAAAUGGCAAAAAGUUAGAUAGUGCGAGGAGGAGGGCUGGGCU .((((.(((.......))).))))........((((((....))))))...(((..(((.(((..................((((.....)))).......(.....)))).)))..))) ( -28.30) >DroSim_CAF1 155215 120 + 1 GCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAAAUGGCAAAAAGUUAGAUAGUGCGAGGAGGAGGGCUGGGCU .((((.(((.......))).))))........((((((....))))))...(((..(((.(((..................((((.....)))).......(.....)))).)))..))) ( -28.30) >DroEre_CAF1 162885 113 + 1 GCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAAAUGGCAAAAAGGUAGAUAGUGCGGACCG---GGCUG---- .....(((((((.((............(((((((((((....))))))....(.....))))))...................((......))........)).))))---)))..---- ( -29.70) >DroYak_CAF1 167859 114 + 1 GCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAAAUGGCAAAAAGUUAGAUAGUGCGGGGCG---GGCAGG--- ((.((((((.(..(.............(((((((((((....))))))....(.....)))))).................((((.....))))....)..).)))))---)))...--- ( -31.30) >consensus GCGUCGUUCGGUGUGAGGAAGAUGAAGUGGGCGCCAGGAAGUCCUGGCAAAAGUUAAGCGCUCAAAUAAAAUAAAGGAAAAUGGCAAAAAGUUAGAUAGUGCGGGGAG___GGCUGG___ .((((.(((.......))).))))...(((((((((((....))))))....(.....)))))).................(.(((.............))).)................ (-23.18 = -22.78 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:23:03 2006