
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,276,053 – 17,276,316 |
| Length | 263 |
| Max. P | 0.997381 |

| Location | 17,276,053 – 17,276,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276053 120 + 23771897 CGUGGGUGGCCUGAACGUGAACGUGAACUUUUCACGUGCGCAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCACCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUU .(((((((......(((....)))(((((....(((.((((.....)))).)))....)))))...........)))))))......(((((((((...........))))))))).... ( -29.80) >DroSec_CAF1 159395 120 + 1 CGUGGGUGGCCUGAACGUGAACGUGAACUUUUCACGUGCGCAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUU .(.(((..((.((((..(((((((((.....))))))((((.....)))))))..))))))..))).).(((((.(....)))))).(((((((((...........))))))))).... ( -30.20) >DroSim_CAF1 154737 120 + 1 CGUGGGUGGCCUGAACGUGAACGUGAACUUUUCACGUGCGCAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUU .(.(((..((.((((..(((((((((.....))))))((((.....)))))))..))))))..))).).(((((.(....)))))).(((((((((...........))))))))).... ( -30.20) >DroEre_CAF1 162390 120 + 1 CGUGGGUGGCCGGAACGUGAACGUGAACUUUUCACGUGCGCAAUUUGUGUUUGUAUUUAGCUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUU ....(((((((((((.(((.((((((.....)))))).))).......(((.......)))........))))).)).)))).....(((((((((...........))))))))).... ( -31.40) >DroYak_CAF1 167348 120 + 1 CGUGGGUGUCCUGAACGUGAACGUGAACUUUUCACGUGCGCAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGUUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUU ...(((.(..(((((..(((((((((.....))))))((((.....)))))))..)))))..).)))......((((((....))))))(((((((...........)))))))...... ( -29.80) >consensus CGUGGGUGGCCUGAACGUGAACGUGAACUUUUCACGUGCGCAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUU .(.(((.(((.((((..(((((((((.....))))))((((.....)))))))..))))))).))).).(((((.(....)))))).(((((((((...........))))))))).... (-28.00 = -28.12 + 0.12)


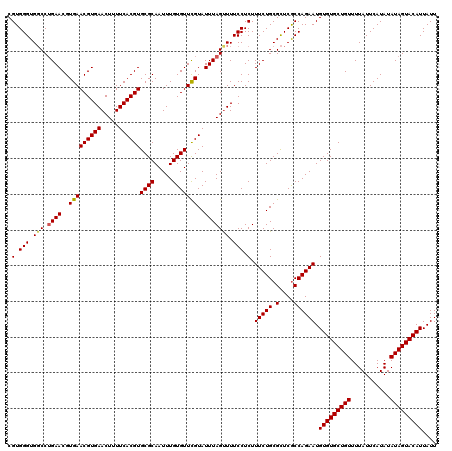
| Location | 17,276,093 – 17,276,205 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -27.14 |
| Energy contribution | -26.78 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276093 112 + 23771897 CAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCACCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCAACCUUUUGCACUGGGUGCUGGGCGA-------- ......(((..((((...((.......))...))))..)))..(((((((((((((...........)))))))).)))))((((..((((........))))...))))..-------- ( -30.00) >DroSec_CAF1 159435 117 + 1 CAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUUUCAACUGGGUUCUGGGCUA--C-CCCA ......(((..((((...((.......))...))))..)))..(((((((((((((...........)))))))).)))))((((.(((((........)))))..))))..--.-.... ( -29.30) >DroSim_CAF1 154777 112 + 1 CAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUUUCAACUGGGUGCUGGGCGA-------- ......(((..((((...((.......))...))))..)))..(((((((((((((...........)))))))).)))))((((.(((((........)))).).))))..-------- ( -29.70) >DroEre_CAF1 162430 109 + 1 CAAUUUGUGUUUGUAUUUAGCUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUU-UGCACUGGGUGCUGGGU---------- ......(((..((((...((.......))...))))..)))..(((((((((((((...........)))))))).)))))((((.(((((.-......)))).).))))---------- ( -23.80) >DroYak_CAF1 167388 120 + 1 CAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGUUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUCUGCACUGGGCGCUGGGUGCUGGGUUCU ......(((..((((...((.......))...))))..)))..(((((((((((((...........)))))))).)))))((((.(((((..(((.....)))..)))).).))))... ( -31.30) >consensus CAAUUUGUGUUCGUAUUUAGUUUUCCUCUUUCUGCGCUCGCCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUUUGCACUGGGUGCUGGGCGA________ ......(((..((((...((.......))...))))..)))..(((((((((((((...........)))))))).)))))((((..((((........))))...)))).......... (-27.14 = -26.78 + -0.36)



| Location | 17,276,133 – 17,276,236 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.74 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276133 103 + 23771897 CCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCAACCUUUUGCACUGGGUGCUGGGCGA-----------------GAACUACCCUUGUCCUAGCCGAACUCAACUC ...(((((((((((((...........)))))))).))))).................((((((((((((((-----------------(.......)))).))))))...))))).... ( -27.90) >DroSec_CAF1 159475 117 + 1 CCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUUUCAACUGGGUUCUGGGCUA--C-CCCAAGGAACAAGGAACUACCCUUGUCCUAGCCGAACUCAACUC ...(((((((((((((...........)))))))).))))).................(((((((((((...--.-))))((((.(((((......)))))))))....))))))).... ( -35.30) >DroSim_CAF1 154817 103 + 1 CCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUUUCAACUGGGUGCUGGGCGA-----------------GAACUACCCUUGUCCUAGCCGAACUCAACUC ...(((((((((((((...........)))))))).)))))....((((((........))))(((((((((-----------------(.......)))).)))))))).......... ( -28.10) >DroEre_CAF1 162470 102 + 1 CCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUU-UGCACUGGGUGCUGGGU-----------------AGGAACUACCCUUGUCCCGGCCGAACUCAACUC ...(((((((((((((...........)))))))).)))))(((..(((...-.((((...)))).((((-----------------((...))))))..)))..)))............ ( -29.20) >DroYak_CAF1 167428 117 + 1 CCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUCUGCACUGGGCGCUGGGUGCUGGGUUCUGGG---AAGGAACUACCCUUGUCCUAGCCGAACUCAACUC ...(((((((((((((...........)))))))).)))))((((((.(((........)))...)))).))(((((((((((---((((......)))..)))))...))))))).... ( -35.40) >consensus CCAGAAUGUGUGCUGUUUUAUUCAUAUAUAGUACAUUAUUCGCCCCGACCUUUUGCACUGGGUGCUGGGCGA_______________AGGAACUACCCUUGUCCUAGCCGAACUCAACUC ...(((((((((((((...........)))))))).)))))((((..((((........))))...)))).................................................. (-21.78 = -21.74 + -0.04)


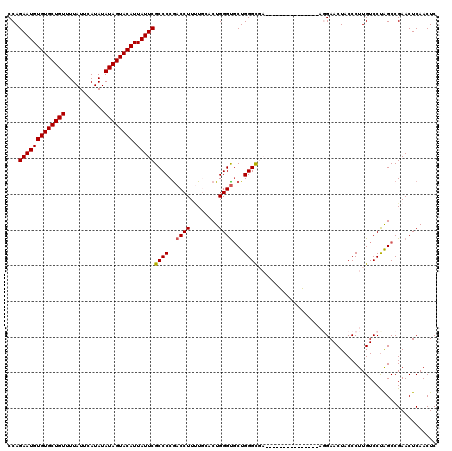
| Location | 17,276,173 – 17,276,276 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276173 103 + 23771897 CGCCCCAACCUUUUGCACUGGGUGCUGGGCGA-----------------GAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUA .((((((...........)))).)).((((((-----------------(.......)))))))..((((((((...(((...(((((....)))))...)))...)))).))))..... ( -31.30) >DroSec_CAF1 159515 117 + 1 CGCCCCGACCUUUUCAACUGGGUUCUGGGCUA--C-CCCAAGGAACAAGGAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUA .((((.(((((........)))))..))))..--.-....((((.(((((......))))))))).((((((((...(((...(((((....)))))...)))...)))).))))..... ( -39.20) >DroSim_CAF1 154857 103 + 1 CGCCCCGACCUUUUCAACUGGGUGCUGGGCGA-----------------GAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUA .(((((............((((((((((((((-----------------(.......)))).))))))...)))))(((((.............)))))))))).((((......)))). ( -31.22) >DroEre_CAF1 162510 102 + 1 CGCCCCGACCUU-UGCACUGGGUGCUGGGU-----------------AGGAACUACCCUUGUCCCGGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUA .(((..(((...-......(((....((((-----------------((...))))))....)))((((..(((((..................)))))..))))..)))..)))..... ( -33.57) >DroYak_CAF1 167468 117 + 1 CGCCCCGACCUUCUGCACUGGGCGCUGGGUGCUGGGUUCUGGG---AAGGAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUA (((((.(..........).))))).(((((((((((..(((((---((((......)))..))))))))(..((((.((((.............))))))))..)......))))))))) ( -36.42) >consensus CGCCCCGACCUUUUGCACUGGGUGCUGGGCGA_______________AGGAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUA .((((..((((........))))...))))....................................((((((((...(((...(((((....)))))...)))...)))).))))..... (-25.74 = -25.70 + -0.04)

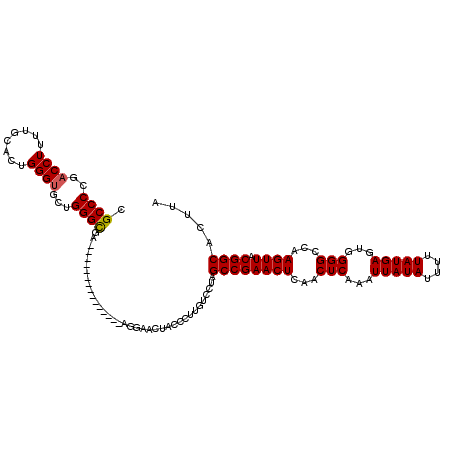
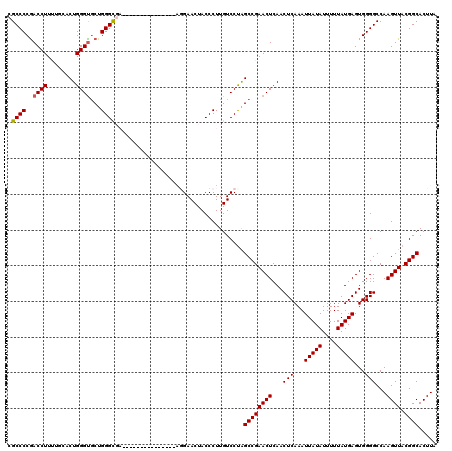
| Location | 17,276,173 – 17,276,276 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276173 103 - 23771897 UAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUC-----------------UCGCCCAGCACCCAGUGCAAAAGGUUGGGGCG .....(((.((((((((((..(((((....((......))....)))))..)))).......((((.....-----------------..)))).((((...))))...)))))).))). ( -31.50) >DroSec_CAF1 159515 117 - 1 UAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUCCUUGUUCCUUGGG-G--UAGCCCAGAACCCAGUUGAAAAGGUCGGGGCG (((((......)))))((((((((((.............)))))....(((((((.(((((((((....))))))))).(((((-.--...))))).....)))))))......))))). ( -41.02) >DroSim_CAF1 154857 103 - 1 UAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUC-----------------UCGCCCAGCACCCAGUUGAAAAGGUCGGGGCG (((((......)))))((((((((((.............)))))....(((((((.((....((((.....-----------------..))))....)).)))))))......))))). ( -31.62) >DroEre_CAF1 162510 102 - 1 UAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCCGGGACAAGGGUAGUUCCU-----------------ACCCAGCACCCAGUGCA-AAGGUCGGGGCG (((((......)))))((((((((((....((......))....)))))..((((.......((((((...))-----------------)))).((((...)))).-..))))))))). ( -37.20) >DroYak_CAF1 167468 117 - 1 UAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUCCUU---CCCAGAACCCAGCACCCAGCGCCCAGUGCAGAAGGUCGGGGCG (((((......)))))((((((((..................((((.((((((..(((((.......).)))).---.)).)))).)))).....((((...))))....))).))))). ( -33.90) >consensus UAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUCCU_______________UCGCCCAGCACCCAGUGCAAAAGGUCGGGGCG .((.((((.(..(((((((..(((((....((......))....)))))..)))))))...).)))).))....................((((...(((..........)))..)))). (-27.86 = -27.78 + -0.08)

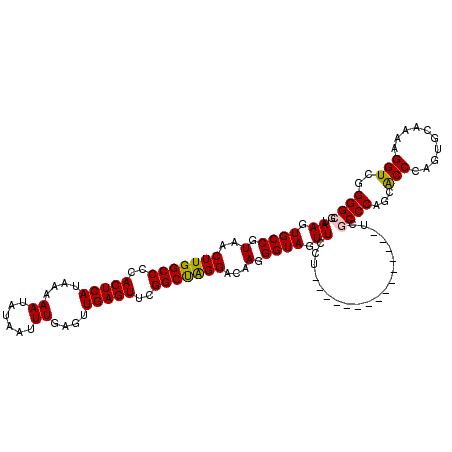
| Location | 17,276,205 – 17,276,316 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -22.76 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276205 111 + 23771897 ---------GAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUAAUGAUGUUGCUUAGCAAAUUGUAGCAACUAAACACAAACA ---------.........((((....((((((((...(((...(((((....)))))...)))...)))).))))..........((((((..((.....)))))))).....))))... ( -23.10) >DroSec_CAF1 159552 120 + 1 AGGAACAAGGAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUAAUGAUGUUGCUUAGCAAAUUGAAGCAACUAAACACAAACA ((((.(((((......))))))))).((((((((...(((...(((((....)))))...)))...)))).))))..........(((((((.........)))))))............ ( -33.30) >DroSim_CAF1 154889 111 + 1 ---------GAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUAAUGAUGUUGCUUAGCAAAUUGAAGCAACUAAACACAAACA ---------.........((((....((((((((...(((...(((((....)))))...)))...)))).))))..........(((((((.........))))))).....))))... ( -23.30) >DroEre_CAF1 162539 113 + 1 -------AGGAACUACCCUUGUCCCGGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUAAUGAUGUUGCUUAGCAAAUUGAAGCAACUAAACACAAACA -------.((.((.......)).)).((((((((...(((...(((((....)))))...)))...)))).))))..........(((((((.........)))))))............ ( -24.60) >DroYak_CAF1 167508 117 + 1 GGG---AAGGAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUAAUGAUGUUGCUUAGCAAAUUGAAGCAACUAAACACAAACA (((---((((......)))).)))..((((((((...(((...(((((....)))))...)))...)))).))))..........(((((((.........)))))))............ ( -28.30) >consensus _______AGGAACUACCCUUGUCCUAGCCGAACUCAACUCAAAUUAUAUUUUUAUGAGUGGGGCCAAGUUACGGCACUUAAUGAUGUUGCUUAGCAAAUUGAAGCAACUAAACACAAACA ..................((((....((((((((...(((...(((((....)))))...)))...)))).))))..........(((((((.........))))))).....))))... (-22.76 = -22.96 + 0.20)

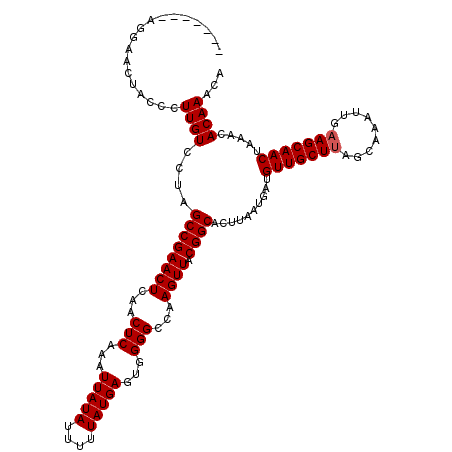

| Location | 17,276,205 – 17,276,316 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17276205 111 - 23771897 UGUUUGUGUUUAGUUGCUACAAUUUGCUAAGCAACAUCAUUAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUC--------- ((....((((((((...........))))))))....))..((.((((.(..(((((((..(((((....((......))....)))))..)))))))...).)))).)).--------- ( -26.90) >DroSec_CAF1 159552 120 - 1 UGUUUGUGUUUAGUUGCUUCAAUUUGCUAAGCAACAUCAUUAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUCCUUGUUCCU ((....((((((((...........))))))))....))......((((.((((..((.....(((.............)))))..))))))))..(((((((((....))))))))).. ( -32.42) >DroSim_CAF1 154889 111 - 1 UGUUUGUGUUUAGUUGCUUCAAUUUGCUAAGCAACAUCAUUAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUC--------- ((....((((((((...........))))))))....))..((.((((.(..(((((((..(((((....((......))....)))))..)))))))...).)))).)).--------- ( -26.90) >DroEre_CAF1 162539 113 - 1 UGUUUGUGUUUAGUUGCUUCAAUUUGCUAAGCAACAUCAUUAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCCGGGACAAGGGUAGUUCCU------- ((....((((((((...........))))))))....))..((.((((.(..(((((((..(((((....((......))....)))))..)))))))...).)))).))...------- ( -28.40) >DroYak_CAF1 167508 117 - 1 UGUUUGUGUUUAGUUGCUUCAAUUUGCUAAGCAACAUCAUUAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUCCUU---CCC ((....((((((((...........))))))))....))..((.((((.(..(((((((..(((((....((......))....)))))..)))))))...).)))).))....---... ( -26.90) >consensus UGUUUGUGUUUAGUUGCUUCAAUUUGCUAAGCAACAUCAUUAAGUGCCGUAACUUGGCCCCACUCAUAAAAAUAUAAUUUGAGUUGAGUUCGGCUAGGACAAGGGUAGUUCCU_______ ((....((((((((...........))))))))....))..((.((((.(..(((((((..(((((....((......))....)))))..)))))))...).)))).)).......... (-27.52 = -27.20 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:56 2006