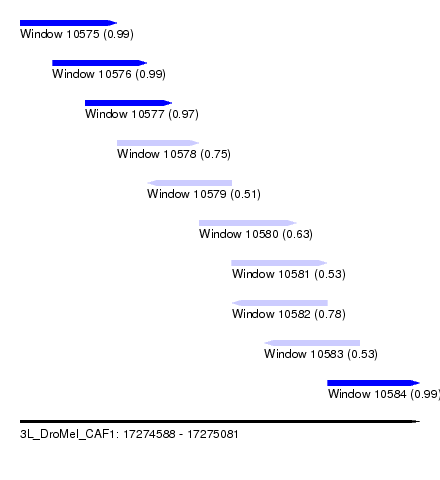
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,274,588 – 17,275,081 |
| Length | 493 |
| Max. P | 0.994008 |

| Location | 17,274,588 – 17,274,708 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -34.12 |
| Energy contribution | -34.52 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274588 120 + 23771897 CCCGAGCGUGUGACGUUCAACAGAAGCAUUCAAAUUAAACUUUAUUUGCCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCU ....(((((((((...((((((((..(...(((((........)))))....(((((.....))))).)..)))).((..(((((........)))))..))....))))))))).)))) ( -36.30) >DroSec_CAF1 158191 120 + 1 CCCGAGCGUGUGACGUUCAACAGAAGCAUUCAAAUUAAACUUUAUUUGCCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCU ....(((((((((...((((((((..(...(((((........)))))....(((((.....))))).)..)))).((..(((((........)))))..))....))))))))).)))) ( -36.30) >DroSim_CAF1 153253 120 + 1 CCCGAGCGUGUGACGUUCAACAGAAGCAUUCAAAUUAAACUUUAUUUGUCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCU ....(((((((((...((((((((..(...(((((........)))))....(((((.....))))).)..)))).((..(((((........)))))..))....))))))))).)))) ( -36.50) >DroEre_CAF1 160980 120 + 1 CCCGAGCGUGUGACGUUCAACAGAAGCAUUCAAAUUAAACUUUAUUUGCCCUCCCCGUACCUUGGGGCGUCCCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCU ....(((((((((...(((((((..((...(((((........)))))....(((((.....))))).))..))).((..(((((........)))))..))....))))))))).)))) ( -33.80) >DroYak_CAF1 165907 120 + 1 CCCGAGCGUGUGACGUUCAACAGAAGCAUUCAAAUUAAACUUUAUUUGCCCUCCCCGUGCCUUUGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCU ....(((((((((...((((((((..(....((((........))))((((.............)))))..)))).((..(((((........)))))..))....))))))))).)))) ( -33.82) >consensus CCCGAGCGUGUGACGUUCAACAGAAGCAUUCAAAUUAAACUUUAUUUGCCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCU ....(((((((((...((((((((.((...(((((........)))))....(((((.....))))).)).)))).((..(((((........)))))..))....))))))))).)))) (-34.12 = -34.52 + 0.40)


| Location | 17,274,628 – 17,274,745 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274628 117 + 23771897 UUUAUUUGCCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAA---AGGAGCAGGAGUUGCGAA .......((((.(((((.....))))).......))))..(((((........)))))((((((.((((.((.(.(((((.((........)).)))))---.))).))))..)))))). ( -37.60) >DroSec_CAF1 158231 117 + 1 UUUAUUUGCCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAA---AGGAGCAGGAGUUGCGAA .......((((.(((((.....))))).......))))..(((((........)))))((((((.((((.((.(.(((((.((........)).)))))---.))).))))..)))))). ( -37.60) >DroSim_CAF1 153293 117 + 1 UUUAUUUGUCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAA---AGGAGCAGGAGUUGCGAA ........(((((((((.....))))).((..((..(((.(((((........)))))((((((((.((....))..)))))))).........)))..---))..))))))........ ( -36.80) >DroEre_CAF1 161020 111 + 1 UUUAUUUGCCCUCCCCGUACCUUGGGGCGUCCCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAA---AGGAGCAGGUCC------ .......(((..(((((.....))))).(..(((..(((.(((((........)))))((((((((.((....))..)))))))).........)))..---)))..).)))..------ ( -36.90) >DroYak_CAF1 165947 120 + 1 UUUAUUUGCCCUCCCCGUGCCUUUGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCACGGGAGGAGCAGGAGUAGCGAA ....(((((((((((.(((((..((..((((....)))..(((((........)))))((((((((.((....))..))))))))..)..))..))))))))))).)))))......... ( -49.80) >consensus UUUAUUUGCCCUCCCCGUACCUUGGGGCGUCUCUGGGCUUUCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAA___AGGAGCAGGAGUUGCGAA ....(((((((((((((.....))))).(((..((..(..(((((........)))))((((((((.((....))..))))))))..)..))..))).....))).)))))......... (-35.24 = -35.28 + 0.04)


| Location | 17,274,668 – 17,274,776 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 97.53 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -35.81 |
| Energy contribution | -35.81 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274668 108 + 23771897 UCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAAGGAGCAGGUCCUGGAACAGUAGCUCCAGCA (((((........)))))((((((((.((....))..))))))))..........((...(((((((((.((((.......)))).)))).........))))).)). ( -36.20) >DroSec_CAF1 158271 108 + 1 UCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAUCGAGCAGGUCCUGGAACAGGACCUCCAGCA (((((........))))).(((..((((((((...(((((.((........)).))))).(.(((....))).)).)))))))((((((((...))))))))...))) ( -37.80) >DroSim_CAF1 153333 108 + 1 UCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAUCGAGCAGGUCCUGGAACAGGACCUCCAGCA (((((........))))).(((..((((((((...(((((.((........)).))))).(.(((....))).)).)))))))((((((((...))))))))...))) ( -37.80) >consensus UCUGCUUAAAAUUGCAGAUUGCAGGCUUGAUCACAUUGCUUGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAUCGAGCAGGUCCUGGAACAGGACCUCCAGCA (((((........)))))((((((((.((....))..))))))))......(((((........(((((.((((.......)))).)))))...........))))). (-35.81 = -35.81 + -0.00)


| Location | 17,274,708 – 17,274,809 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274708 101 + 23771897 UGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAAGGAGCAGGUCCUGGAACAGUAGCUCCAGCAGCAGUAGCUGGAACAGGAGCAGUCAGACAAACA ...........((((((.......(((((.((((.......)))).)))))........(((((..((((....)))).....))))).)))).))..... ( -31.80) >DroSec_CAF1 158311 101 + 1 UGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAUCGAGCAGGUCCUGGAACAGGACCUCCAGCAGCAGUAGCUGGAACAGGAGCAGUCAGACAAACA ...........((((((...(.(((....))).).......((.(((((((...)))))))((((((.......))))))......)).)))).))..... ( -29.60) >DroSim_CAF1 153373 101 + 1 UGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAUCGAGCAGGUCCUGGAACAGGACCUCCAGCAGCAGUAGCUGGAACAGGAGCAGUCAGACAAACA ...........((((((...(.(((....))).).......((.(((((((...)))))))((((((.......))))))......)).)))).))..... ( -29.60) >consensus UGCAAAUGAACGUUGGCAAAGGAGCAGGAGUUGCGAAUCGAGCAGGUCCUGGAACAGGACCUCCAGCAGCAGUAGCUGGAACAGGAGCAGUCAGACAAACA ...........((((((.......(((((.((((.......)))).)))))..........((((((.......)))))).........)))).))..... (-28.20 = -28.20 + -0.00)


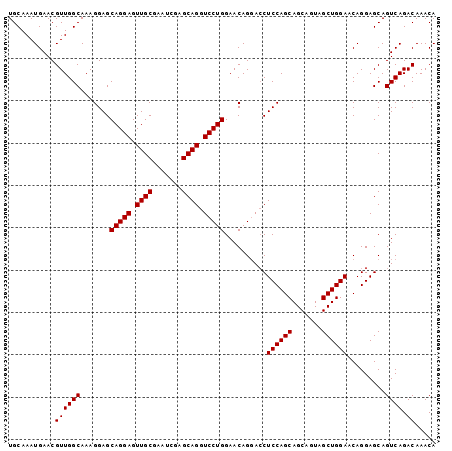
| Location | 17,274,745 – 17,274,849 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274745 104 - 23771897 ACAAAAAAUCUCCUUUAUUGCAUGCACAGCACGUUUCAUGUGUUUGUCUGACUGCUCCUGUUCCAGCUACUGCUGCUGGAGCUACUGUUCCAGGACCUGCUCCU ...................(((.....(((((((...))))))).((((....((....((((((((.......))))))))....))....)))).))).... ( -25.90) >DroSec_CAF1 158348 104 - 1 ACAAAAAAUCUCCUUUAUUGCAUGCACAGCACGUUUCAUGUGUUUGUCUGACUGCUCCUGUUCCAGCUACUGCUGCUGGAGGUCCUGUUCCAGGACCUGCUCGA ....................((.(((.(((((((...)))))))))).))...((......((((((.......))))))(((((((...))))))).)).... ( -29.60) >DroSim_CAF1 153410 104 - 1 ACAAAAAAUCUCCUUUAUUGCAUGCACAGCACGUUUCAUGUGUUUGUCUGACUGCUCCUGUUCCAGCUACUGCUGCUGGAGGUCCUGUUCCAGGACCUGCUCGA ....................((.(((.(((((((...)))))))))).))...((......((((((.......))))))(((((((...))))))).)).... ( -29.60) >consensus ACAAAAAAUCUCCUUUAUUGCAUGCACAGCACGUUUCAUGUGUUUGUCUGACUGCUCCUGUUCCAGCUACUGCUGCUGGAGGUCCUGUUCCAGGACCUGCUCGA ....................((.(((.(((((((...)))))))))).))...((......((((((.......))))))(((((((...))))))).)).... (-25.20 = -25.87 + 0.67)


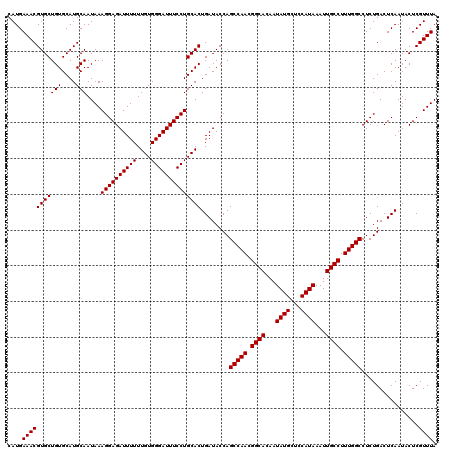
| Location | 17,274,809 – 17,274,929 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274809 120 + 23771897 CAUGAAACGUGCUGUGCAUGCAAUAAAGGAGAUUUUUUGUGGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUA ....(((((((((((....)))....((((((((((....))))))))))))))........(((((.((((...((((...))))...)))).)))))................)))). ( -30.50) >DroSec_CAF1 158412 120 + 1 CAUGAAACGUGCUGUGCAUGCAAUAAAGGAGAUUUUUUGUGGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUA ....(((((((((((....)))....((((((((((....))))))))))))))........(((((.((((...((((...))))...)))).)))))................)))). ( -30.50) >DroSim_CAF1 153474 120 + 1 CAUGAAACGUGCUGUGCAUGCAAUAAAGGAGAUUUUUUGUGGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUA ....(((((((((((....)))....((((((((((....))))))))))))))........(((((.((((...((((...))))...)))).)))))................)))). ( -30.50) >DroEre_CAF1 161207 120 + 1 CAUGAAACGUGCUGUGCAUGCAAUAAAGGAGAUUUUUUGUGGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUA ....(((((((((((....)))....((((((((((....))))))))))))))........(((((.((((...((((...))))...)))).)))))................)))). ( -30.50) >DroYak_CAF1 166138 120 + 1 CAUGAAACGUGCUGUGCAUGCAAUAAAGGAGAUUUUUUGUGGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUA ....(((((((((((....)))....((((((((((....))))))))))))))........(((((.((((...((((...))))...)))).)))))................)))). ( -30.50) >consensus CAUGAAACGUGCUGUGCAUGCAAUAAAGGAGAUUUUUUGUGGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUA ....(((((((((((....)))....((((((((((....))))))))))))))........(((((.((((...((((...))))...)))).)))))................)))). (-30.50 = -30.50 + 0.00)

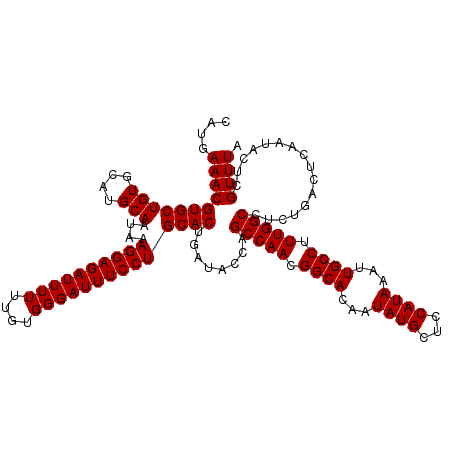
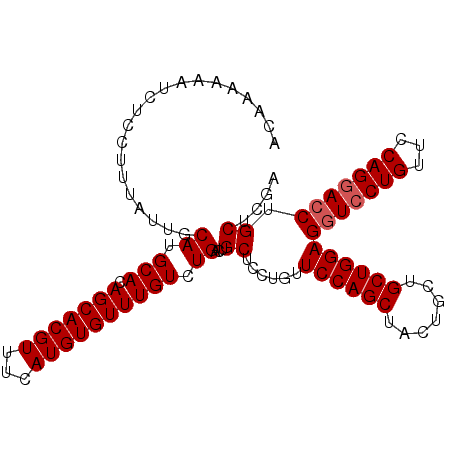
| Location | 17,274,849 – 17,274,967 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274849 118 + 23771897 GGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUAU--AUGUAUGUAUACCUUCACAUGGAUAUUCCAGUUGCCA (((....)))((((((......(((((.((((...((((...))))...)))).)))))....................((--((.(((((........))))).))))..))).))).. ( -26.40) >DroSec_CAF1 158452 120 + 1 GGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUAUAUAUGUAUGUAUACCUUCACAGUGAUAUUUCAGUUGCCA (((....)))(((((((.....(((((.((((...((((...))))...)))).)))))((.(((....((((.(((......)))...))))....))).)).......)))).))).. ( -26.50) >DroSim_CAF1 153514 120 + 1 GGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUAUAUAUGUAUGUAUGCCUUCACAGUGAUAUUUCAGUUGCCA (((....)))(((((((.....(((((.((((...((((...))))...)))).)))))((.(((....((((.(((......)))...))))....))).)).......)))).))).. ( -27.60) >DroEre_CAF1 161247 113 + 1 GGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUAU------AUGUAUACCUUCACAUGGACAUUUCAG-AGCCA (((....)))((.((((.....(((((.((((...((((...))))...)))).)))))................((((((------.((........)).))))))...))))-.)).. ( -26.40) >DroYak_CAF1 166178 114 + 1 GGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUAU------AUGUAUACCUUCACAUGGAUAUUUCAGGAGCCA .((...(((((....((((((((((((.((((...((((...))))...)))).)))))...(((....((((........------..))))....)))..))).)))).))))).)). ( -27.60) >consensus GGGAUUUCCUGCACUGAUACCAGCCAACGGCACAAUAUGCUCCAUAAAUUGCCUUUGGCCUCUGACUCAAUACUCGUUUAU__AUGUAUGUAUACCUUCACAUGGAUAUUUCAGUUGCCA (((....)))(((((((.....(((((.((((...((((...))))...)))).)))))...(((....((((................))))....)))..........)))).))).. (-20.65 = -21.25 + 0.60)


| Location | 17,274,849 – 17,274,967 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -27.05 |
| Energy contribution | -27.13 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274849 118 - 23771897 UGGCAACUGGAAUAUCCAUGUGAAGGUAUACAUACAU--AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGAGCAUAUUGUGCCGUUGGCUGGUAUCAGUGCAGGAAAUCCC .(....).(((...((((((((...(....)...)))--))......(((((((......((((((.((((...((((...))))...)))).))))))....))))))).)))..))). ( -33.70) >DroSec_CAF1 158452 120 - 1 UGGCAACUGAAAUAUCACUGUGAAGGUAUACAUACAUAUAUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGAGCAUAUUGUGCCGUUGGCUGGUAUCAGUGCAGGAAAUCCC ..(((.((((..(((((((.(((....((((................))))....))).))(((((.((((...((((...))))...)))).))))))))))))))))).((....)). ( -32.79) >DroSim_CAF1 153514 120 - 1 UGGCAACUGAAAUAUCACUGUGAAGGCAUACAUACAUAUAUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGAGCAUAUUGUGCCGUUGGCUGGUAUCAGUGCAGGAAAUCCC ..(((.((((..(((((((.(((...(((((................))).))..))).))(((((.((((...((((...))))...)))).))))))))))))))))).((....)). ( -32.19) >DroEre_CAF1 161247 113 - 1 UGGCU-CUGAAAUGUCCAUGUGAAGGUAUACAU------AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGAGCAUAUUGUGCCGUUGGCUGGUAUCAGUGCAGGAAAUCCC .....-........((((((((........)))------))......(((((((......((((((.((((...((((...))))...)))).))))))....))))))).)))...... ( -31.00) >DroYak_CAF1 166178 114 - 1 UGGCUCCUGAAAUAUCCAUGUGAAGGUAUACAU------AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGAGCAUAUUGUGCCGUUGGCUGGUAUCAGUGCAGGAAAUCCC .((.((((.........(((((........)))------))......(((((((......((((((.((((...((((...))))...)))).))))))....)))))))))))...)). ( -31.90) >consensus UGGCAACUGAAAUAUCCAUGUGAAGGUAUACAUACAU__AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGAGCAUAUUGUGCCGUUGGCUGGUAUCAGUGCAGGAAAUCCC ..(((.((((..((((..(.(((....((((................))))....))).)((((((.((((...((((...))))...)))).))))))))))))))))).((....)). (-27.05 = -27.13 + 0.08)



| Location | 17,274,889 – 17,275,007 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274889 118 - 23771897 UAAAACUAAAUCAUAGACAAGUAUAAACUAUUGUCUGGGGUGGCAACUGGAAUAUCCAUGUGAAGGUAUACAUACAU--AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGA ..........(((((((..(((....))).((((((....((((..((((...((.(.(((....((((......))--))..))).).))....))))..)))).))))))))))))). ( -22.10) >DroSec_CAF1 158492 120 - 1 UAAAACUAAAUCAUAGACAAGUAUAAACUAUUGUCUGGGGUGGCAACUGAAAUAUCACUGUGAAGGUAUACAUACAUAUAUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGA ..........(((((((..(((....))).((((((....((((..((((..((..(((((....(((((......)))))..)).)))..))..))))..)))).))))))))))))). ( -26.50) >DroSim_CAF1 153554 120 - 1 UAAAACUAAAUCAUAGACAAGUAUAAACUAUUGUCUGGGGUGGCAACUGAAAUAUCACUGUGAAGGCAUACAUACAUAUAUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGA ..........(((((((..(((....))).((((((....((((..((((....(((.((((....)))).((((............))))))).))))..)))).))))))))))))). ( -23.50) >DroEre_CAF1 161287 113 - 1 UAAAACUAAAUCAUAGACAAGUAUAAACUAUUGUCUGGGGUGGCU-CUGAAAUGUCCAUGUGAAGGUAUACAU------AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGA ..........(((((((..(((....))).((((((..(((..((-((((..(((..(((((........)))------))..))).........)))))))))..))))))))))))). ( -23.50) >DroYak_CAF1 166218 114 - 1 CAAAACUAAAUCAUAGACAAGUAUAAACUAUUGUCUGGGGUGGCUCCUGAAAUAUCCAUGUGAAGGUAUACAU------AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGA ..........(((((((..(((....))).((((((....((((..((((((((..((((((........)))------))....)..))))...))))..)))).))))))))))))). ( -24.60) >consensus UAAAACUAAAUCAUAGACAAGUAUAAACUAUUGUCUGGGGUGGCAACUGAAAUAUCCAUGUGAAGGUAUACAUACAU__AUAAACGAGUAUUGAGUCAGAGGCCAAAGGCAAUUUAUGGA ..........(((((((..(((....))).((((((....((((..((((.(((((........)))))...............(((...)))..))))..)))).))))))))))))). (-20.10 = -20.02 + -0.08)


| Location | 17,274,967 – 17,275,081 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -19.36 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17274967 114 + 23771897 CCCCAGACAAUAGUUUAUACUUGUCUAUGAUUUAGUUUUAGCGCCCAAAUAGGGAUGCAGCUGA-AAAGGAAACACAUCCUUUCUAUACUCUU-----ACCCAACUAACCAAAAUAAUAA ....(((((..(((....)))))))).....(((((((((((.(((.....))).....)))))-((((((......))))))..........-----....))))))............ ( -21.30) >DroSec_CAF1 158572 114 + 1 CCCCAGACAAUAGUUUAUACUUGUCUAUGAUUUAGUUUUAGCGCCCAAAUAGGGAUGCAGCUGA-AAAGGACACACAUCCUUUCUAUACCCUU-----ACCCAACUAACCAAAAUAAUAA ....(((((..(((....)))))))).....(((((((((((.(((.....))).....)))))-((((((......))))))..........-----....))))))............ ( -21.10) >DroSim_CAF1 153634 115 + 1 CCCCAGACAAUAGUUUAUACUUGUCUAUGAUUUAGUUUUAGCGCCCAAAUAGGGAUGCAGCUCAAAAAGGACACACAUCCUUUCUAUACCCUU-----ACCCAACUAACCAAAAUAAUAA ....(((((..(((....)))))))).....((((((...((((((.....))).))).......((((((......))))))..........-----....))))))............ ( -20.00) >DroEre_CAF1 161360 119 + 1 CCCCAGACAAUAGUUUAUACUUGUCUAUGAUUUAGUUUUAGCGCCCAAAAAGGGAUGCAGCUGA-AAAGGACACACUUCCUUUCCGUACCCAAAAUUUACCGAACUAACAAAAAUAAUAA ....(((((..(((....)))))))).....(((((((..((((((.....))).))).((.((-((.(((......))))))).))..............)))))))............ ( -22.70) >DroYak_CAF1 166292 114 + 1 CCCCAGACAAUAGUUUAUACUUGUCUAUGAUUUAGUUUUGGCGCUCUAAAGGGGAUGCAGCUGG-AAAGGACACCCUUUCUUUCUAUACCCUA-----ACCCAGCUGACCAAAAUAAUAA ....(((((..(((....))))))))........(((((((((((.....((((.((.((..((-(((((....)))))))..)).)))))).-----....))).).)))))))..... ( -29.80) >consensus CCCCAGACAAUAGUUUAUACUUGUCUAUGAUUUAGUUUUAGCGCCCAAAUAGGGAUGCAGCUGA_AAAGGACACACAUCCUUUCUAUACCCUU_____ACCCAACUAACCAAAAUAAUAA ....(((((..(((....)))))))).....((((((...((((((.....))).))).......((((((......))))))...................))))))............ (-19.36 = -18.72 + -0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:50 2006